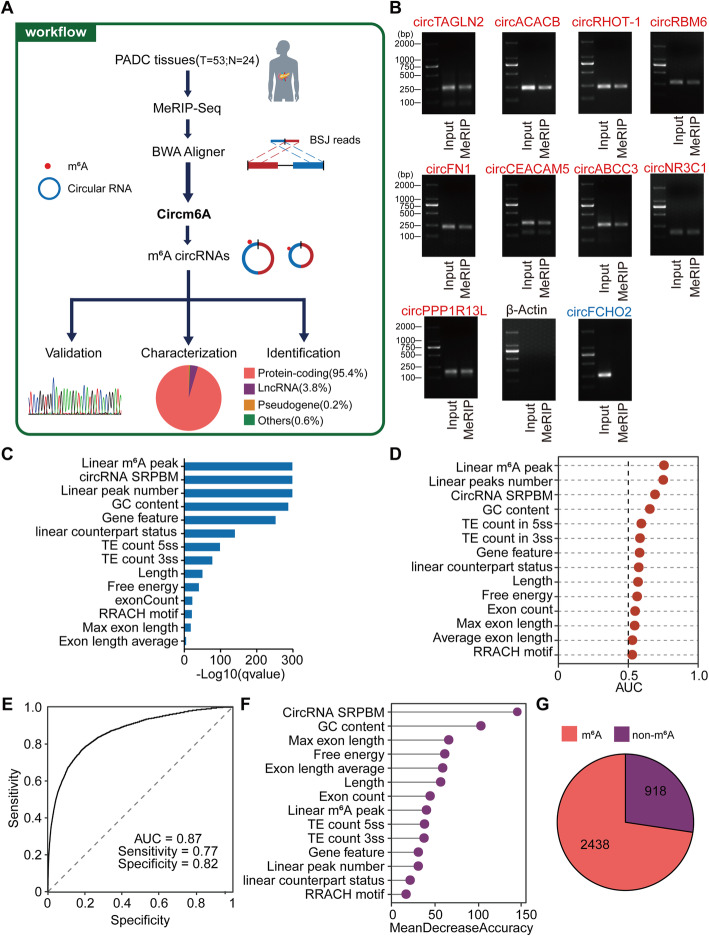
Fig. 3.
Identification and characterization of m6A-circRNAs in PDAC. A Workflow for the identification and characterization of m6A-circRNAs from MeRIP-seq of 77 tissue samples from 53 PDAC patients using Circm6A. B Validate the m6A modification of 9 m6A-circRNAs using MeRIP-qPCR after RNase R treatment. C Differential analysis results for 14 features for predicting m6A-circRNAs. Bars show FDR-corrected P values (q value). P values for quantitative features were calculated with the Wilcoxon rank sum test, and P values for qualitative variables were calculated with Pearson’s chi-squared test. D The power of prediction of m6A-circRNAs for the 14 features. E Receiver operating characteristic (ROC) curve for the random forest model in ten-fold cross-validation. F Mean decrease accuracy (MDA) for each feature in the random forest model. G The predicted result for the m6A status of “ambiguous circRNAs” by the random forest model. m6A, “ambiguous circRNAs” predicted as m6A-circRNAs; non-m6A, “ambiguous circRNAs” predicted as non-m6A circRNAs