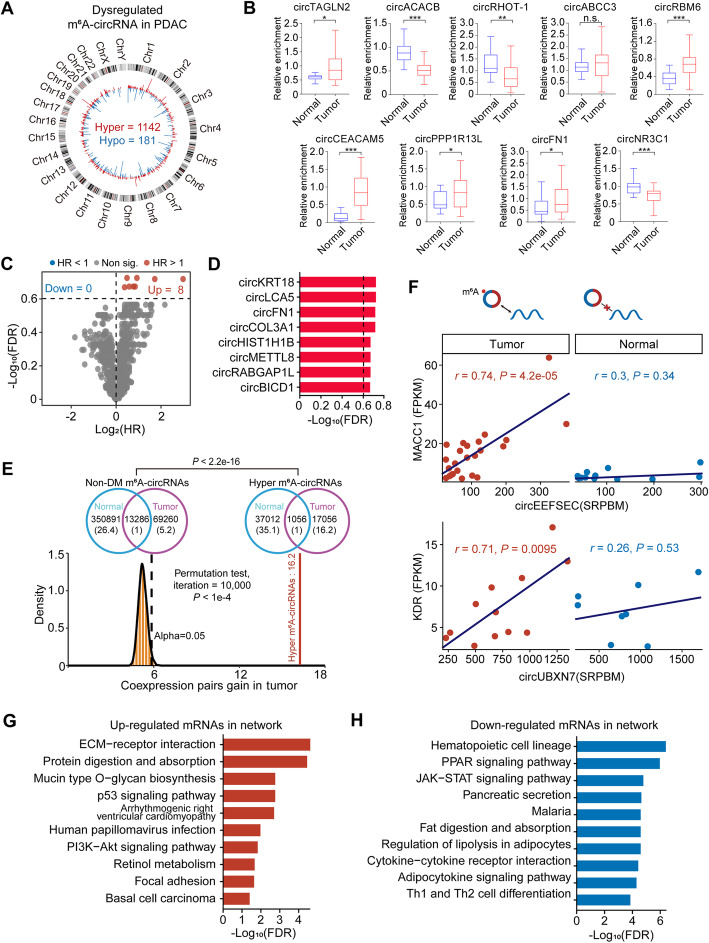
Fig. 4.
Differential m6A methylation of m6A-circRNAs between PDAC tumor and normal tissues. A The circos plot shows the genome-wide hypermethylated (red bar) and hypomethylated (blue bar) circRNAs in PDAC tumor tissues. The height of the bar represents the frequency of m6A modification in all samples. B Validation of differentially methylated m6A-circRNAs using MeRIP-qPCR. *, P < 0.05; **, P < 0.01; ***, P < 0.001; n.s. P > 0.05 of Student’s t test. C, D The correlation between PFS and m6A level of differentially methylated m6A-circRNAs in PDAC. Volcano plot depicting log2 hazard ratios (HRs) and − log10 (FDR) values of differentially methylated m6A-circRNAs in the Cox proportional hazards model of PFS. The horizontal dashed line in C and vertical dashed line in D correspond to “FDR = 0.25”. All 8 differentially methylated m6A-circRNAs significantly associated with PFS are showed in D. E The number of coexpressed pairs for hypermethylated m6A-circRNAs and non-differentially methylated m6A-circRNAs (non-DM m6A-circRNAs) in PDAC tumor and normal tissue samples, respectively (upper panel). The number in parentheses is normalized to the number of coexpressed pairs shared between the tumor and normal samples. The permutation test shows significant contribution of hypermethylated m6A-circRNAs to the gain of coexpression network in tumor tissue samples compared with randomly selected non-DM m6A-circRNAs (lower panel). Vertical red lines indicate the gain fold of hypermethylated m6A-circRNAs, the histogram indicates the fold gain of randomly selected non-DM m6A-circRNAs. F Scatter plots showing the coexpression of circEEFSEC-MACC1 pair (upper panel) and circUBXN7-KDR pair (lower panel) in PDAC tumor and normal tissue samples, respectively. r represents Pearson correlation coefficients; P represents P value. G, H Top enriched KEGG pathways for upregulated mRNAs (red, G) and downregulated mRNAs (blue, H) in the coexpression network that hypermethylated m6A-circRNAs were involved in