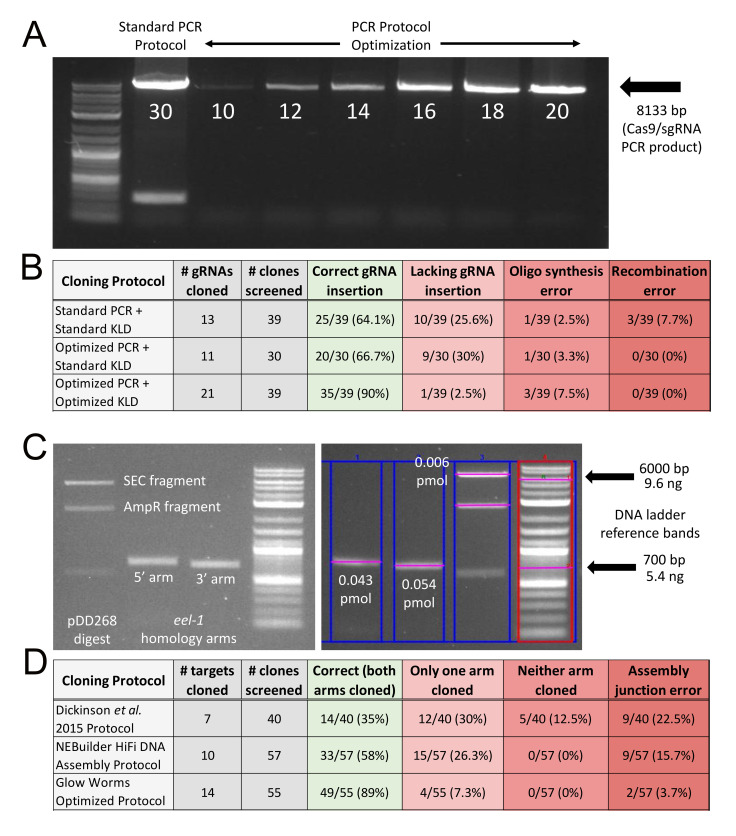
Figure 1. Highly improved cloning efficiency for plasmid-based CRISPR knock-in.
(A) Lane 1: NEB® DNA ladder standard (N3200S); Lane 2: A standard NEB® Q5 PCR protocol of 30 cycles results in too much DNA based on light smearing and an additional unwanted PCR product of ~300bp that reproducibly appears with 30 cycles; Lanes 3-8: Optimizing PCR cycle number for pDD162 amplification. Based on this data, we chose 15 cycles as the optimal number for all subsequent amplifications of pDD162. (B) Too much PCR product and insufficient DpnI digestion results in ~35% of KLD ligation clones being incorrect Cas9/sgRNA plasmids. In contrast, optimizing both the PCR and KLD ligation reactions results in 90% of clones having a correct gRNA insertion. (C) Case study of DNA yields for the vector backbone and homology arms used for construction of an FP/SEC repair plasmid, in this case for the gene eel-1. Left panel: post clean-up electrophoresis of a pDD268 SpeI/ClaI digestion and eel-1 homology arm PCRs. Right panel: Another gel electrophoresis of the same pDD268 digestion and eel-1 homology arms, this time with DNA quantification using Bio-Rad Image Lab 6.0 software to measure the intensity of ethidium bromide staining relative to an NEB® DNA ladder standard (N3200S). The 6000bp band of the ladder was used to determine the number of ng of SEC fragment and the 700bp band for the number of ng of each homology arm. Number of pmol was calculated as described in Methods. (D) Thorough quantification of DNA and subsequent optimization of the amount of DNA in the Gibson assembly reaction dramatically improves cloning efficiency for the FP/SEC plasmid, reproducibly reaching 89%. The “Glow Worms Optimized Protocol” is detailed in Methods.