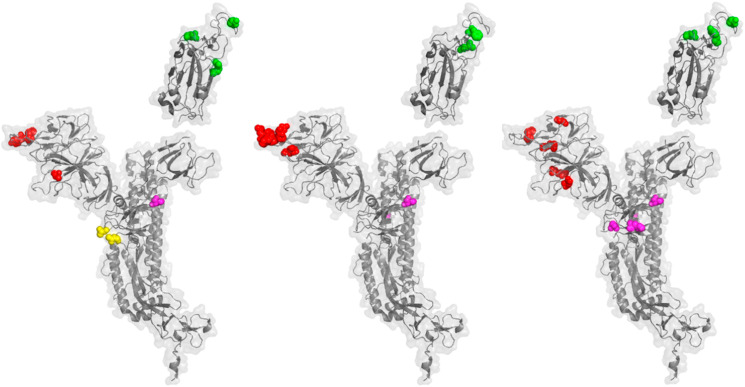
Fig. 3.
Emerging variants of interest exhibit distinguishing mutations in the same patterns as variants of concern. A. A ribbon diagram and transparent surface are shown for the SARS-CoV-2 spike protein (PDB 7CWN) with spheres indicating sites of mutation that distinguish the Mu variant, B.1.621. Red spheres indicate mutations in the NTD; T95I, Y144T, Y144S. Green spheres indicate mutations in the RBD; E484K, N501Y. Magenta spheres indicate an interchain contact mutation; D614G. Yellow spheres indicate a mutation in the fusion cleavage site; P681H. B. The SARS-CoV-2 spike protein is shown with spheres indication the positions mutations that distinguish the lambda variant, C.37. Red spheres; G75V, T76I, 246–253 RSYLTPGD. Green spheres; L452Q, F490S. Magenta spheres indicate interchain contact mutations; D614G, T859 N. C. The SARS-CoV-2 spike protein is shown with spheres indication the positions mutations that distinguish the variant of interest C.1.2. Red spheres; C136F, R190S, D215G, Δ144, Δ242-243. Green spheres; Y449H, E484K, N501Y. Magenta spheres indicate interchain contact mutations; D614G, H655Y, N679K, T859 N. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)