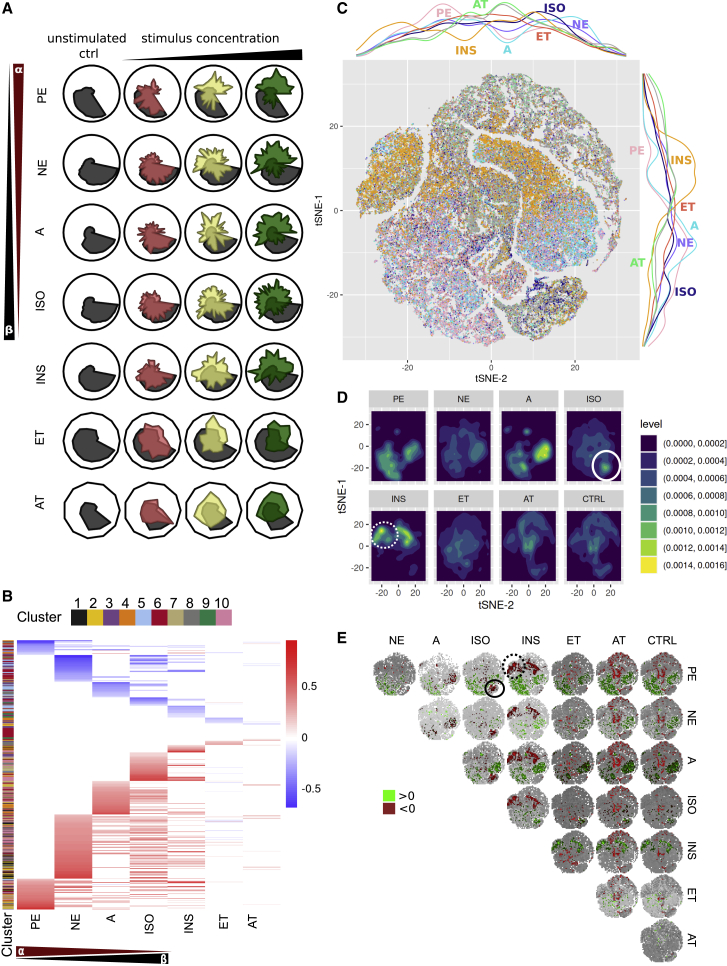
Figure 3.
Population-level and single-cell phenotyping of canonical hypertrophic stimuli
(A) Radar plots show meta-feature values for the canonical hypertrophic stimuli aggregated on a population level (three concentrations: red, low; yellow, intermediate; green, high; untreated control, gray).
(B) Comparative analysis of the features selected for the canonical hypertrophic stimuli. For each stimulus, the dose-dependent dynamic is color coded (red indicates an increase and blue a decrease; clusters were calculated on an unstimulated control). See also Figure S4D.
(C) Single-cell phenotyping of the canonical hypertrophic stimuli: t-distributed stochastic neighbor embedding (t-SNE) of all single cells measured in one experiment (highest substance concentrations, substances are color coded). Marginal densities for each stimulus over the t-SNE plot are shown by colored lines to the right and at the top of the t-SNE plot.
(D) Densities per stimulus on the t-SNE plots from (C). Visually, areas of notably higher density represent subpopulations of potential interest and are circled for ISO and for INS are indicated with a dotted line.
(E) Quantification of stimulus-induced differences, pairwise differences of binned and normalized data (50 × 50 grid, rate differences were calculated: x − y axis). The most extreme differences are colored in red (upper 5%) and green (lowest 5%). Quantitatively identified substance-specific subpopulations are circled for ISO and for INS are circled with a dotted line.
In (A) and (B), data are shown for 5 independent experiments. In (C)–(E), single-cell analysis of 1 experiment is shown.