Abstract
Therapeutic interventions aimed at inducing fetal hemoglobin and reducing the concentration of sickle hemoglobin is an effective approach to ameliorating acute and chronic complications of sickle cell disease, exemplified by the long-term use of hydroxyurea. However, there remains an unmet need for the development of additional safe and effective drugs for single agent or combination therapy for individuals with β-hemoglobinopathies. Regulation of the γ-globin to β-globin switch is achieved by chromatin remodeling at the HBB locus on chromosome 11 and interactions of major DNA binding proteins, such as KLF1 and BCL11A in the proximal promoters of the globin genes. Experimental evidence also supports a role of epigenetic modifications including DNA methylation, histone acetylation/methylation, and microRNA expression in γ-globin gene silencing during development. In this review, we will critically evaluate the role of epigenetic mechanisms in γ-globin gene regulation and discuss data generated in tissue culture, pre-clinical animal models, and clinical trials to support drug development to date. The question remains whether modulation of epigenetic pathways will produce sufficient efficacy and specificity for fetal hemoglobin induction and to what extent targeting these pathways form the basis of prospects for clinical therapy.
Keywords: Epigenetics, sickle cell disease, fetal hemoglobin, DNA methylation, histone acetylation, microRNA
Impact statement
Therapeutic options to induce fetal hemoglobin that are safe and effective are a high priority for treating individuals with sickle cell disease. Tissue culture, preclinical, and clinical studies provide evidence for targeting epigenetic mechanisms of reversing the γ-globin to β-globin gene switch during development. The goal of this review is to critically assess the role of epigenetic mechanisms in γ-globin gene regulation and discuss the question whether modulation of epigenetic pathways will produce sufficient efficacy and specificity for fetal hemoglobin induction to warrant future development for clinical therapy.
Introduction
Sickle cell disease (SCD) is one of the most common inherited blood disorders that affect more than 100,000 individuals in the United States and millions of people worldwide. 1 Fetal hemoglobin (HbF) is the major genetic modulator of the hematologic and clinical manifestations of SCD. For example, people with hereditary persistence of fetal hemoglobin maintain high HbF levels of 20–30% and have less symptoms and better clinical outcomes. 2 Thus, therapeutic interventions aimed at inducing HbF expression while reducing the concentration of sickle hemoglobin (HbS), are effective approaches to ameliorating complications of SCD. To achieve this goal, additional knowledge about the molecular and cell signaling mechanisms involved in regulating γ-globin transcription are required. Induction of HbF requires chromatin remodeling of histones at the γ-globin (HBG) and β-globin gene promoters.3,4 Epigenetic modifications including acetylation and methylation influence access of trans-acting DNA-binding proteins such as GATA1, TAL1, E2A, LMO2, LRF/ZBTB7, and LDB1, which facilitate DNA looping of the locus control region (LCR) with each globin promoter, to enhance gene transcription.5,6 Pre-clinical and clinical evidence described to date suggests that epigenetic modifications mediated by DNA methyltransferases (DNMTs), histone methyltransferases, histone demethylases, histone deacetylases (HDACs), and histone acetyltransferases (HATs) along with changing microRNA (miRNA) expression levels, may be involved in the γ-globin to β-globin switch. However, it remains unclear how important these epigenetic mechanisms are in HbF induction and whether drugs that target changes in epigenetic marks will provide sufficient efficacy and potency to achieve clinical utility. In this review, we will critically assess epigenetic mechanisms involved in globin gene regulation and discuss the question as to whether modulation of epigenetic pathways will produce sufficient efficacy and specificity for HbF induction to warrant expanded development for clinical therapy.
Food and drug administration-approved therapies for sickle cell disease
Disease-targeted treatment options for individuals with SCD were limited to hydroxyurea (HU) for over two decades. More recently, three additional agents were FDA-approved including Oxbryta® to inhibit HbS polymerization, Crizulizamab® to block interactions with P-selectin glycoprotein ligand 1 (PSGL-1), and Endari® to increase L-glutamine levels and reduce oxidative stress within sickle cells.7–9 After many decades of research, no other HbF-inducing agents have been FDA approved, creating an unmet need for additional drug development for SCD.
Epigenetic mechanisms of globin gene regulation
A broad spectrum of pharmaceutical agents has demonstrated the ability to induce HbF in tissue culture; however, their efficacy in preclinical animal models and clinical studies has not been demonstrated. Many of these molecules act independently of DNA sequence-based genetic mutations of the γ-globin gene. Experimental data suggest these mechanisms, termed epigenetics, including DNA methylation, histone acetylation/methylation and changing microRNA (miRNA) expression levels,10–17 alter transcription of the γ-globin gene resulting in reversal of the γ- to β-globin switch.10,18–21
DNA methylation
DNA methylation was the first described epigenetic signal postulated to play a role in globin gene regulation during development.22–28 In 1980, van der Ploeg et al. 23 analyzed the methylation status of the γ-globin promoter by digestion of DNA isolated from erythroid and nonerythroid tissues, with methylation sensitive restriction enzymes and Southern blot analysis. They were among the first group to establish a significant correlation between human γ-globin promoter methylation levels and gene transcription during development. 23 Shortly after, Busslinger et al. 28 showed that methylation of nucleotides located at positions −760 to +100 in the 5ʹ region of the γ-globin gene prevented gene transcription. 28 Early studies by Comi et al. in 198629 showed that treatment of adult erythroid cells with bromo-deoxyuridine, a nucleotide analog that inhibits DNA methylation, reactivated HbF expression. 29
Since the discovery of the role of DNA methylation in globin gene regulation, a large body of literature supports changes in methylation levels of the human HBB locus on chromosome 11, in contributing to hemoglobin switching mechanisms. 14 ,18,19,30–34 During developmentally regulated erythroid cell differentiation, the five globin genes (5-ʹε, Gγ, Aγ, δ, β-3ʹ) in the HBB locus become sequentially demethylated and transcriptionally active. 35 Specifically, in late gestation, the CpG dinucleotides of the γ-globin promoters are hypomethylated supporting gene expression until after birth with completion of hemoglobin switching by one year of age in normal infants.32, 36 This finding was further supported by Singh et al. 37 showing that γ-globin promoter hypermethylation in early fetal liver gradually demethylates as erythroid differentiation progressed over time. 37 Conversely, during adult erythropoiesis, the γ-globin gene promoter becomes hypermethylated and silenced by DNA methylation machinery,18,30–34 producing a 20-fold repression in gene transcription. 36
To achieve epigenetic regulation, DNMTs interact with various transcription factors that bind the γ-globin proximal promoter facilitating highly methylated CpG sites located at positions −256, −162, −71, −53, −50, +6, +17, and +5010,38 Additionally, chromatin regulators, including DNMT3A, the lysine methyltransferase SUV4-20 h1, the serine/threonine kinase CK2alpha, and components of the Nucleosome Remodeling and Deacetylase (NuRD) co-repressor complex, occupy the γ-globin promoter to repress its expression.39,40 It has been shown that silencing of DNMT1 in primary cultures of erythroid progenitor cells derived from CD34+ stem cells isolated from baboon bone marrow cells, increased levels of γ-globin mRNA and decreased DNA methylation of the proximal promoter. 41 These findings highlight the involvement of DNA methylation machinery and cofactors in regulating chromatin structure in the HBB locus and γ-globin gene silencing in adult-stage erythroid cells.
Transcriptional repressors of γ-globin gene expression
Genetic mapping and later genome-wide association studies (GWAS) discovered quantitative trait loci in the B-cell lymphoma/leukemia 11 A (BCL11A) gene, an XmnI variant upstream of the Gγ-globin gene, and the HBS1L-MYB intergenic region that were associated with inherited variations in human HbF levels.42–46 The C > T genetic variant in XmnI (rs7482144) located at −158 upstream of the Gγ-globin gene was correlated with disease severity in adults with β-thalassemia. 43 The BCL11A gene encodes a C2H2 type zinc-finger protein, which is a major transcriptional repressor of γ-globin during adult erythropoiesis.47,48 Inherited polymorphisms in BCL11A associated with HbF levels in hemoglobinopathy patients 49 provided the impetus for laboratory studies demonstrating BCL11A transcription factor as a major silencer of γ-globin gene expression (Figure 1). KLF1 was the first major transcription factor discovered to play a key role in adult β-globin gene activation during hemoglobin switching. 50 Later studies demonstrated the role of KLF1 in the γ- to β-globin gene switching process by directly activating BCL11A, 51 which indirectly repressed γ-globin expression.52–55 KLF1 also regulates many components of the cell cycle machinery during erythropoiesis.52,54,56,57
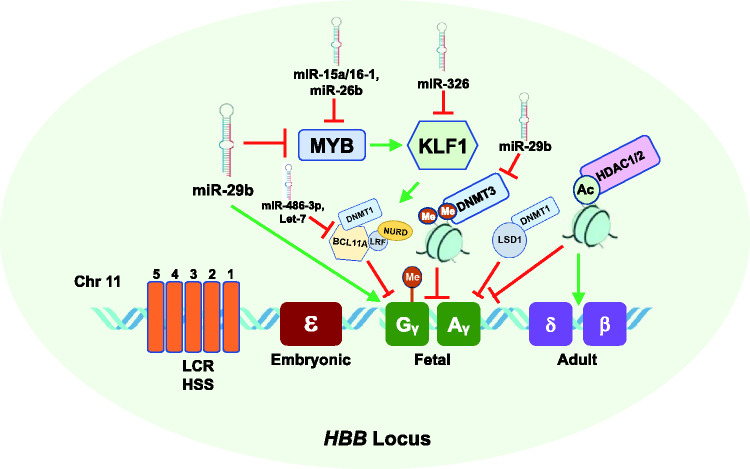
Figure 1.
Effect of epigenetic mechanisms, miRNAs, and transcription factors on fetal globin gene expression during hemoglobin switching. Shown is the HBB gene locus on chromosome 11 which consists of the five developmentally regulated globin genes, including fetal γ-globin (green) and adult β-globin (purple). Silencing of γ-globin genes is mediated by transcriptional repressors MYB which activates KLF1, which in turn activates the repressor BCL11A which mediates transcriptional silencing of γ-globin. The γ-globin promoters become hypermethylated and silenced by DNMT1. The DNMT3 proteins, DNMT3A and DNMT3B, are required for long-term methylation of the γ-globin gene promoters and silencing during adult erythropoiesis. In addition, deacetylation by HDAC1/2 inhibits γ-globin expression and activates adult HBB expression. MiR-29b, a DNMT3 inhibitor, inhibits MYB expression resulting in γ-globin gene activation. Additional miRNAs that inhibit (red line) or induce (green line) HbF are shown. (A color version of this figure is available in the online journal.)
DNMT: DNA methyltransferase; BCL11A; B-cell lymphoma 11A; HSs: hypersensitive sites, KLF1; Krüppel-like factor 1; LCR; locus control region; miR: microRNA.
Additional studies support an indirect role of the oncogene MYB 58 in regulating γ-globin transcription through targeting BCL11A and the repressor TR2/TR4.38,54 MYB is highly expressed in immature hematopoietic cells and down regulated during erythropoiesis. 59 Overexpression of MYB transcription factor inhibits γ-globin expression in K562 cells, 58 and MYB silencing induces HbF expression in primary human erythroid progenitors. 60 In human genetic studies, the association of MYB with HbF levels was demonstrated using quantitative trait loci studies and subsequent functional assays.43,58,60 The leukemia/lymphoma-related factor (LRF), encoded by the ZBTB7A gene, is a ZBTB transcription factor that directly binds fetal γ-globin promoter and represses γ-globin gene expression during adult erythropoiesis through a NuRD repressor complex independent of BCL11A 61 (Figure 1).
Ten eleven-translocation dioxygenases
The TET enzymes are a family of TET methylcytosine dioxygenases that are involved in DNA demethylation. 62 We previously showed that the γ-globin gene transcriptional activator, NRF2, which modulated chromatin structure in the human HBB locus of β-YAC transgenic mice during development, interacted with TET3 to regulate γ-globin methylation. 16 Tet2 and Tet3 dioxygenases that catalyze formation of 5-hydroxymethylcytosine (5 hmC) and are expressed during early stages of erythroid differentiation were also shown to regulate γ-globin expression by mediating demethylation of the γ-globin promoter. 18 Conversely, Sp1 binding to the −71 to −34 sequence of γ-globin increased upon site-specific cytosine methylation, suggesting Sp1 is a potential repressor of γ-globin expression.63,64
Protein methyltransferases and demethylases
Another group of epigenetic modifiers include lysine demethylases (LSD), which are enzymes that can demethylate mono- and di-methylated lysines. 65 They are known to interact with BCL11A to influence γ-globin gene silencing in both murine and human adult erythroid cells. 66 Specifically, LSD1 associates with BCL11A through a complex containing the repressor element-1 silencing factor co-repressor-1 (CoREST) to mediate γ-globin gene silencing. 66 Supporting studies using RNA interference to silence LSD1 or the LSD1 inhibitor tranylcypromine support a role for LSD1 in γ-globin regulation.66,67 A study by Rivers et al. 68 demonstrated that oral administration of the LSD1 inhibitor ORY-3001 increased F-reticulocytes, γ-globin mRNA, and HbF synthesis in baboons and SCD mice. 68 However, there were concerns with potential adverse effects when tested in preclinical transgenic mice and primate models34, 67,68 (Table 1).
Table 1.
Summary of epigenetic therapeutics tested in clinical trials for sickle cell disease.
| Drug | Mechanism of action | Dates | Phase | Subject number |
Age, yrs | Number sites |
ClinicalTrials.gov identifier (status) |
|---|---|---|---|---|---|---|---|
| Decitabine | DNMT1 inhibitor | 6/2011–2/2016 | 2 | 10 | >18 | 1 |
NCT01375608 (completed) |
| Decitabine/THU | DNMT1 inhibitor/cytidine deaminase inhibitor | 8/2012–6/2018 | 1 | 25 | >18 | 1 |
NCT01685515 (completed) |
| Nicotinamide, THU, Decitabine |
vitB3 analogue, cytidine deaminase inhibitor, DNMT1 inhibitor | 1/2020–3/2021 | 1 | 20 | >18 | 1 |
NCT04055818 (recruiting) |
| Vorinostat | HDAC inhibitor | 10/2009–10/2014 | 2 | 15 | >18 | 3 |
NCT01000155 (terminated) |
| Panobinostat | HDAC inhibitor | 11/2010–7/2024 | 1 | 18 | >18 | 1 |
NCT01245179 (active, not recruiting) |
| Arginine butyrate | HDAC inhibitor | 9/1997–2/2005 | 2 | 23 | 16–60 | 4 |
NCT00004412 (completed) |
| INCB059872 | LSD1 inhibitor | 4/2017–10/2018 | 1 | 12 | >18 | 8 |
NCT03132324 (terminated) |
| HQK-1001 | SCFA/HDAC inhibitor | 4/2011–3/2012 | 2 | 52 | 12–60 | 16 |
NCT01322269 (completed) |
| HQK-1001 | SCFA/HDAC inhibitor | 7/2012–12/2013 | 2 | 77 | 12–60 | 18 |
NCT01601340 (terminated) |
| HQK-1001 | SCFA/HDAC inhibitor | 3/2009–5/2010 | 1 | 21 | 12–60β-thal | 2 |
NCT00790127 (completed) |
| HQK-1001 | SCFA/HDAC inhibitor | 3/2009–7/2010 | 2 | 24 | 12–60 | 9 |
NCT00842088 (completed) |
| HQK-1001 | SCFA/HDAC inhibitor | 5/2012–1/2013 | 2 | 10 | 16–50β-thal | 1 |
NCT01642758 (completed) |
| HQK-1001 | SCFA/HDAC inhibitor | 3/2012–12/2012 | 2 | 10 | 18–55β-thal | 1 |
NCT01609595 (completed) |
HDAC: histone deacetylase; DNMT1: DNA methyltransferase 1; LSD1: lysine-specific histone demethylase 1; SCFA: short chain fatty acid; THU: tetrahydrouridine.
Another group of proteins involved in globin gene regulation is the methylated DNA binding protein (MBD) family. The MBD family includes methyl CpG binding protein 2 (MeCP2), which interacts with DNA containing methylated CpG-rich sequences with high affinity. 69 MBD proteins regulate gene activity by modifying chromatin through recruitment of histone deacetylases and methylases to methylated DNA. 69 Our group and others have shown that MeCP2 and/or other MBD proteins are involved in γ-globin regulation in adult erythroid progenitors.15,63,70 Indeed, depletion of MBD2 produced a 10 to 20-fold increase in γ-globin expression in β-globin yeast artificial chromosome transgenic mice.71,72 Mechanistically, proteins in the MBD family recruit chromatin remodeling members of the NuRD co-repressor complex. The latter is an important epigenetic reader of DNA methylation that regulates gene transcription during normal development and neoplastic diseases15,70 through protein–protein interactions. Studies in primary adult erythroid cells showed that knockdown of the MBD2-NuRD complex increased the γ/γ + β mRNA ratio by 10-fold which correlated with an increase in HbF protein levels. 72 Other studies showed that the direct repeat erythroid-definitive (DRED) and BCL11A-NuRD co-repressor complexes, containing DNMT1 protein, were also involved in γ-globin gene silencing in adult erythroid cells.66,73
Collectively, these studies provide strong evidence of a critical role for DNA methylation and DNA methylation machinery in hemoglobin switching during adult erythropoiesis and highlight the potential of DNA methylation-targeted therapies for HbF induction. However, studies have shown that HbF levels of ∼30% with pancellular distribution are required to ameliorate SCD clinical severity. 74 Therefore, it is unclear as to what extent the increase in γ-globin transcription produced by repression of the MBD2-NuRD complex translates into clinical efficacy.
DNA methylation agents in HbF induction
Two DNMT inhibitor agents, namely 5-azacytidine and its analog decitabine, have been investigated for many years as HbF-inducing agents75–83 (Table 1). 5-azacytidine was first demonstrated to induce HbF in anemic baboons 84 and later in individuals with SCD and β-thalassemia.85–90 Decitabine and 5-azacytidine are both DNMT inhibitors that reactivate γ-globin expression via hypomethylation of the CpG site within the γ-globin gene promoter which allows binding of erythroid-specific transcription factors such as KLF1, BCL11A, GATA-1, and GATA-2 to chromatin sites in the HBB locus.18,51,91 Indeed, decitabine is a potent HbF inducer when given parenterally but oral administration leads to rapid degradation by intestinal cytidine deaminases.92,93 Furthermore, there are major concerns with adverse side effects, including neutropenia, thrombocytopenia, thrombophilia, or cytotoxicity from taking decitabine.94–96
More recent efforts have focused on the administration of tetrahydrouridine (THU) before decitabine to prevent rapid degradation in the liver 93 (Table 1). In fact, a recent clinical trial combining oral decitabine and THU showed promising results associated with increased HbF and total hemoglobin levels in persons with SCD. 31 In 2020, the US FDA approved the combination of oral cedazuridine, a synthetic nucleoside analog derived from THU, and decitabine (INQOVI) formulation for use in adults with myelodysplastic syndrome. 97 Supporting studies in acute myeloid leukemia patients and in non-human primate baboons showed that pharmacological inhibitors of DNMT, namely S110, 5-aza-2ʹ-deoxycytidine, and decitabine, induced high levels of HbF.18,84,98,99 A newer generation methyltransferase inhibitor, adenosine-2′, 3′ dialdehyde (Adox) was shown to induce γ-globin gene expression in human primary erythroid cells in culture. 100 The recent discovery of a novel orally bioavailable DNMT1-selective inhibitor, GSK3482364 which directly blocks DNMT1 enzymatic activity while sparing DNMT3A or DNMT3B, decreased γ-globin promoter methylation and increased HbF expression in adult human erythroid progenitor cells and SCD transgenic mice. 101 However, due to a limited number of therapies to treat SCD and concerns about adverse effects from decitabine and other DNA methylation inhibitors with known cytotoxic effects,94,95 there remains an unmet need for the development of novel non-cytotoxic agents to target DNA methylation machinery to induce HbF.
Histone modifications
Post translational histone modifications including histone acetylation and methylation are involved in the regulation of γ-globin gene transcription. 102 The developmentally regulated expression of the globin genes requires sequential interaction with the LCR, which consists of five DNA I hypersensitive sites (HSs), with the individual globin promoters. The process of histone acetylation is strongly correlated with an open chromatin conformation and increased gene transcriptional activity, whereas methylation of histone residues can either increase or decrease gene transcription, depending on the location and number of amino acid residues methylated. 102 For instance, formation of histone H3K4me3 is usually associated with transcriptional activation, while histone H3K9me3 and H3K27me3 are associated with inaccessible chromatin and gene silencing.103,104 The general process of histone acetylation is catalyzed by HATs which transfer an acetyl functional group via acetyl coenzyme A from one molecule to another. Increased histone acetylation is typically associated with chromatin opening resulting in the binding of transcription factors to activate gene expression, whereas histone deacetylation tends to condense chromatin into a tightly packed state, which prevents binding of regulatory factors.
Histone acetylation
Indeed, increased histone acetylation has been strongly associated with increased γ-globin gene expression. 13 Supporting studies have implicated the process of acetylation and methylation of lysine residues on histone tails in HbF induction in erythroid cells and preclinical animal models.10,105–108 Specifically, acetylation of H3 and H4 lysine residues and recruitment of RNA polymerase II to the γ-globin promoter region results in elevated HbF expression.12,109 Conversely, deacetylation of the lysine residues within the N-terminal tails of H3 and H4 histones has been observed to shift the balance of γ-globin gene expression resulting in gene silencing.110,111 The observation of ectopic expression of the protein deacetylase SIRT1 and SIRT activators (SRT2104 and SRT1720) increased γ-globin gene expression in primary human erythroid cells and K562 cells was surprising. 112 Interestingly, SIRT1 also increased binding of RNA polymerase II and acetylation of H4K16 at the γ-globin promoter, thereby enhancing gene transcription. 112 Another study reported that histone acetylation is dependent on binding of the erythroid-specific transcriptional activators GATA-1 and NF-E2 at the HBB locus which was not sufficient for γ-globin transcription. 113 However, Layon et al. 114 reported that GATA-1 alone was sufficient to direct chromatin structure reorganization of the HBB LCR and an erythroid pattern of gene expression in the absence of other hematopoietic transcription factors such as NF-E2. 114
The complexity of the relationship between histone deacetylation and globin gene regulation has been investigated in earlier studies by Perrine et al. 108 who demonstrated that the HDAC inhibitor, butyrate, significantly delayed the globin gene switch in sheep fetuses compared with controls. Similar studies found that specific histone acetylation patterns play a critical role in the developmental switch of the murine HBB genes. 106 Their findings, along with other supporting studies,106–108 led to the discovery of butyrate in silencing γ-globin activation in erythroid progenitors, preclinical models, and patients with SCD.106,107,115–117 In addition to butyrate, HDAC1 and HDAC2 have been further recognized as molecular targets mediating HbF induction through a chemical genetic strategy and RNA interference. 118 Moreover, our group showed that butyrate and other HDAC inhibitors, including trichostatin A, alter p38 MAP kinase and STAT-5 signaling leading to γ-globin gene deactivation.119–121
Histone methylation
Histone methylation states can be associated with gene activation, gene silencing, or a bivalent state, to further regulate γ-globin gene expression.122–124 Protein arginine methyltransferase family member 1 (PRMT1), which regulates the action of HATs, was shown to influence γ-globin gene silencing through interactions with FoP. 125 The interaction between PRMT1 and FoP facilitates in the acetylation of Lys9/Lys14 and subsequent transcription of the adult HBB gene. 126 This suggests that PRMT1 indirectly favors activation of HBB gene expression by interacting with repressor proteins and competing for regulatory binding sites in the γ-globin promoters. Similarly PRMT5 was shown to contribute to human γ-globin gene silencing by interacting with the nuclear protein, LYAR (human homologue of mouse Ly-1 antibody reactive clone), and triggering histone H4 Arg3 symmetric demethylation by DNMT3A in adult erythroid progenitors.39,127 PRMT5 was shown to also interact with the histone lysine methyltransferase Suv4-20h1 and components of the NuRD complex to induce additional repressive epigenetic marks at the γ-globin promoter. 40 These data highlight the complexity of the relationship between histone-modifying events and recruitment of factors at the HBB locus that regulate globin gene expression.
Histone modifying agents in HbF induction
A number of HDAC inhibitors aimed at targeting epigenetic silencing of HbF expression in patients with SCD and other β-hemoglobinopathies have been investigated in clinical trials116,117, 128 (Table 1). The HDAC inhibitor, butyrate, has been studied by our laboratory119,120 and others for its role in HbF induction. 129 In 1999, Atweh et al. 117 reported that SCD patients who responded to intermittent arginine butyrate maintained high HbF levels without adverse side effects for a mean of 29.9 weeks, as long as they continued treatment 117 (Table 1). Other butyrate analogues, including phenylbutyrate, isobutyramide and other short chain fatty acids, have also been tested in clinical trials for β-thalassemia due to their capacity to induce HbF in human erythroid cells.130–132 The orally bioavailable short-chain fatty acid derivative sodium 2, 2 dimethyl butyrate (HQK-1001) was tested in the phase I/II trial consisting of 21 adult patients with β-thalassemia intermedia syndromes; however, it was not well tolerated 133 (Table 1). These findings question the potential for butyrate and its analogues as potential HbF inducer agents for treatment of patients with SCD and other β-hemoglobinopathies. Extensive efforts have been made to improve the effectiveness of HDAC inhibitors while decreasing unwanted side effects.
Epigenetic modifiers in clinical trials
A limited number of epigenetic modulators to treat SCD or other β-hemoglobinopathies have been tested in phase I or II clinical trials. Most of these drugs are intended for treatment of different forms of cancer, and as such there are major concerns about off-target effects and cytotoxicity. We summarize trials registered in Clinicaltrials.gov of epigenetic drugs that have been tested but have not proceeded to FDA approval for SCD treatment (Table 1).
MiRNAs and HbF induction
MiRNAs are short (∼22 nucleotides) noncoding RNA molecules that associate with the miRNA-induced silencing complex (mRISC) to silence gene-specific mRNA in the cytoplasm. 134 MiRNAs facilitate mRNA degradation or suppression of translation by base-pairing to complementary sequences in the 3ʹ-untranslated regions of target mRNA.135,136 The human genome encodes over 1000 miRNAs genes, which may target several different mRNAs to inhibit or restore gene expression. 137 Table 2 summarizes miRNAs that have been associated with γ-globin gene activation and HbF induction.
Table 2.
Summary of microRNAs and its target genes in HbF induction.
| miRNA | Target gene |
References |
|---|---|---|
| miR-326 | KLF1 | 138 |
| miR-26B | GATA-1, MYB | 139 , 140 |
| miR-151-3p | BCL11A, MYB | 140 |
| miR-29B | DNMT3A and 3B | 15 |
| miR-144 | NRF2 | 17 |
| miR-486-3p | BCL11A | 141 |
| miR-223 | BCL11A, LMO2 | 142 , 143 |
| miR-210 | BCL11A, raptor | 144 , 145 , 146 |
| miR-15a, miR-16-1 | MYB | 60 , 140 |
| miR34a | STAT3 | 147 |
| miR-23a/27a | KLF3 and SP1 | 148 |
Recent reports highlight the therapeutic potential of miRNAs to ameliorate the severity of hemoglobinopathies, by also targeting globin genes or their regulatory pathways resulting in HbF induction. For instance, a study by Bianchi et al. 144 showed that miR-210 was upregulated in erythroid precursors of a thalassemia patient with high HbF levels. 144 The authors also observed that mithramycin targeted miR-210 as a mechanism of γ-globin activation in K562 cells. 144 In another study investigating SCD patients treated with HU, expression of miR-26b and miR-151-3p was associated with HbF levels at maximum tolerated doses. 149 Previous studies by our group demonstrated that miR-34a mediated HbF induction in K562 cells by silencing STAT3 expression, a known repressor of γ-globin. 147 In subsequent functional studies in normal and sickle erythroid progenitors, we demonstrated that miR-144 silenced NRF2 transcription and concomitant repression of γ-globin transcription, which was reversed in a dose-dependent manner by miR-144 antagomir. 17
This finding is important considering that we, and others, have investigated pharmacological NRF2 activators as a class of promising HbF-inducing agents for the treatment of SCD. 150 For example, Lee et al. 151 showed that overexpression of tumor promoting LIN28B decreased miR-Let7 expression and increased γ-globin gene expression and HbF levels in cultured adult primary erythroid cells, in part via targeting BCL11A expression. 151 Similarly, miR‐486‐3p mediated HbF induction in adult erythroid progenitors by inhibiting BCL11A expression although the role of direct epigenetic changes in the actions of either LIN28B or miR-486-3p remains unknown. 141 Another study demonstrated that miR-15a and miR-16–1 induced HbF by targeting the γ-globin repressor protein MYB in infants with trisomy 13, as a mechanism of delayed switching. 60
Our recently published work demonstrated that miR-29b functions as an HbF inducer in KU812 cells and normal human erythroid progenitors by targeting DNMT3 gene silencing. 15 Interestingly, overexpression of miR-29b also improves common hematological malignancies.152,153 Our published findings on the role of miR29b in HbF induction is the first study to provide evidence of a miRNA-DNA methylation interaction as a mechanism of HbF induction. Future studies to evaluate the ability of miR-29b to induce HbF in vivo are ongoing.
Non-coding small RNAs, such as miR-29b, are attractive molecules for inhibiting repressors of γ-globin gene transcription. However, there are challenges with delivery of miRNAs into cells due to their short half-life under physiological conditions and unfavorable immune response and concerns with potentially undesirable off-target effects. 154 To overcome these challenges, conjugate-mediated drug delivery has been established as a promising platform for safe and targeted small interfering RNA delivery in vivo such as cholesterol conjugation. This modification works by two primary mechanisms: (1) intercalation into the plasma membrane and internalization by endocytosis and (2) binding circulating plasma lipoproteins to promote interactions with lipoprotein receptors.155,156 Several cholesterol-conjugated small interfering RNAs have advanced to clinical evaluation such as RXI-109 for its ability to regulate connective tissue growth in age-related macular degeneration. 157 The novel concept of “miRNA supplement therapy” is currently in clinical trials for the treatment of diseases, such as myelodysplastic syndrome and cancer.158,159 It is believed that approximately 30–50% of all protein-coding genes are possibly regulated by miRNAs in health and deregulated in disease.
Summary
In summary, we highlight epigenetic mechanisms and miRNAs implicated in γ-globin gene transcription and discuss HbF-inducing epigenetic therapeutics as novel treatment approaches for SCD. Current research efforts to expand SCD treatment options support the potential ability of DNA methylation and HDAC inhibitors to be developed as HbF-inducing agents for treatment of SCD. Whether these agents play a direct role in developmentally regulated hemoglobin switching similar to major transcription repressors such as BCL11A remains a question. The discovery of the DNMT inhibitor Decitabine and HDAC inhibitor arginine butyrate for the treatment of SCD has produced sufficient clinical phenotype data and HbF induction to warrant continued investigation. However, there are many challenges associated with the development of these agents for oral administration and long-term impact on disease severity and response to treatment.160,161
Research to establish a direct role for epigenetic mechanisms and miRNAs in HbF induction is at its infancy. Additional studies to demonstrate a role in globin gene regulation and their ability to produce an additive effect on HbF induction in preclinical animal models might justify future clinical trials. The potential to target epigenetic proteins such as DNMT1, HDACs, or miRNAs involved in γ-globin gene silencing provides new therapeutic approaches to reverse globin gene silencing during adult development for the successful treatment of individuals with β-hemoglobinopathies.
Footnotes
AUTHORS’ CONTRIBUTIONS: ASD, AF, and BSP reviewed the literature and shared in writing several drafts of the paper. ASD identified and designed the review sections, figures and tables and conducted final review of paper.
DECLARATION OF CONFLICTING INTERESTS: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
FUNDING: The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This work was supported in part by HL144641-01A1 to AS-D and HL149365-01A1 to BSP.
ORCID iD: Athena Starlard-Davenport https://orcid.org/0000-0002-8938-237X
References
- 1.Weatherall DJ. The role of the inherited disorders of hemoglobin, the first "molecular diseases," in the future of human genetics. Annu Rev Genomics Hum Genet 2013; 14:1–24 [DOI] [PubMed] [Google Scholar]
- 2.Akinsheye I, Alsultan A, Solovieff N, Ngo D, Baldwin CT, Sebastiani P, Chui DH, Steinberg MH. Fetal hemoglobin in sickle cell anemia. Blood 2011; 118:19–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Saunthararajah Y. Targeting sickle cell disease root-cause pathophysiology with small molecules. Haematologica 2019; 104:1720–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gribnau J, Diderich K, Pruzina S, Calzolari R, Fraser P. Intergenic transcription and developmental remodeling of chromatin subdomains in the human beta-globin locus. Mol Cell 2000; 5:377–86 [DOI] [PubMed] [Google Scholar]
- 5.Martyn GE, Wienert B, Yang L, Shah M, Norton LJ, Burdach J, Kurita R, Nakamura Y, Pearson RCM, Funnell APW, Quinlan KGR, Crossley M. Natural regulatory mutations elevate the fetal globin gene via disruption of BCL11A or ZBTB7A binding. Nat Genet 2018; 50:498–503 [DOI] [PubMed] [Google Scholar]
- 6.Liu N, Hargreaves VV, Zhu Q, Kurland JV, Hong J, Kim W, Sher F, Macias-Trevino C, Rogers JM, Kurita R, Nakamura Y, Yuan GC, Bauer DE, Xu J, Bulyk ML, Orkin SH. Direct promoter repression by BCL11A controls the fetal to adult hemoglobin switch. Cell 2018; 173:430–42.e17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.https://www.fda.gov/drugs/resources-information-approved-drugs/fda-disco-l-glutamine-sickle-cell-disease (2017, accessed 15 June 2021)
- 8.https://www.fda.gov/drugs/resources-information-approved-drugs/fda-approves-voxelotor-sickle-cell-disease (2019, accessed 15 June 2021)
- 9.https://www.novartis.com/news/media-releases/new-novartis-medicine-adakveo-crizanlizumab-approved-fda-reduce-frequency-pain-crises-individuals-living-sickle-cell-disease (2019, accessed 15 June 2021)
- 10.Ginder GD. Epigenetic regulation of fetal globin gene expression in adult erythroid cells. Transl Res 2015; 165:115–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ahmadvand M, Noruzinia M, Fard AD, Zohour MM, Tabatabaiefar MA, Soleimani M, Kaviani S, Abroun S, Beiranvand S, Saki N. The role of epigenetics in the induction of fetal hemoglobin: a combination therapy approach. Int J Hematol Oncol Stem Cell Res 2014; 8:9–14 [PMC free article] [PubMed] [Google Scholar]
- 12.Yin W, Barkess G, Fang X, Xiang P, Cao H, Stamatoyannopoulos G, Li Q. Histone acetylation at the human beta-globin locus changes with developmental age. Blood 2007; 110:4101–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fathallah H, Weinberg RS, Galperin Y, Sutton M, Atweh GF. Role of epigenetic modifications in normal globin gene regulation and butyrate-mediated induction of fetal hemoglobin. Blood 2007; 110:3391–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lavelle D, Vaitkus K, Hankewych M, Singh M, DeSimone J. Developmental changes in DNA methylation and covalent histone modifications of chromatin associated with the epsilon-, gamma-, and beta-globin gene promoters in papio anubis. Blood Cells Mol Dis 2006; 36:269–78 [DOI] [PubMed] [Google Scholar]
- 15.Starlard-Davenport A, Smith A, Vu L, Li B, Pace BS. MIR29B mediates epigenetic mechanisms of HBG gene activation. Br J Haematol 2019; 186:91–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhu X, Xi C, Ward A, Takezaki M, Shi H, Peterson KR, Pace BS. NRF2 mediates gamma-globin gene regulation through epigenetic modifications in a beta-YAC transgenic mouse model. Exp Biol Med 2020; 245:1308–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li B, Zhu X, Ward CM, Starlard-Davenport A, Takezaki M, Berry A, Ward A, Wilder C, Neunert C, Kutlar A, Pace BS. MIR-144-mediated NRF2 gene silencing inhibits fetal hemoglobin expression in sickle cell disease. Exp Hematol 2019; 70:85–96 e5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ruiz MA, Rivers A, Ibanez V, Vaitkus K, Mahmud N, DeSimone J, Lavelle D. Hydroxymethylcytosine and demethylation of the gamma-globin gene promoter during erythroid differentiation. Epigenetics 2015; 10:397–407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mabaera R, Richardson CA, Johnson K, Hsu M, Fiering S, Lowrey CH. Developmental- and differentiation-specific patterns of human gamma- and beta-globin promoter DNA methylation. Blood 2007; 110:1343–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Adelvand P, Hamid M, Sardari S. The intrinsic genetic and epigenetic regulator factors as therapeutic targets, and the effect on fetal globin gene expression. Expert Rev Hematol 2018; 11:71–81 [DOI] [PubMed] [Google Scholar]
- 21.Fathallah H, Portnoy G, Atweh GF. Epigenetic analysis of the human alpha- and beta-globin gene clusters. Blood Cells Mol Dis 2008; 40:166–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McGhee JD, Ginder GD. Specific DNA methylation sites in the vicinity of the chicken beta-globin genes. Nature 1979; 280:419–20 [DOI] [PubMed] [Google Scholar]
- 23.van der Ploeg LH, Flavell RA. DNA methylation in the human gamma Delta beta-globin locus in erythroid and nonerythroid tissues. Cell 1980; 19:947–58 [DOI] [PubMed] [Google Scholar]
- 24.Shen CK, Maniatis T. Tissue-specific DNA methylation in a cluster of rabbit beta-like globin genes. Proc Natl Acad Sci U S A 1980; 77:6634–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mavilio F, Giampaolo A, Care A, Migliaccio G, Calandrini M, Russo G, Pagliardi GL, Mastroberardino G, Marinucci M, Peschle C. Molecular mechanisms of human hemoglobin switching: selective undermethylation and expression of globin genes in embryonic, fetal, and adult erythroblasts. Proc Natl Acad Sci U S A 1983; 80:6907–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Enver T, Zhang JW, Papayannopoulou T, Stamatoyannopoulos G. DNA methylation: a secondary event in globin gene switching? Genes Dev 1988; 2:698–706 [DOI] [PubMed] [Google Scholar]
- 27.Perrine SP, Greene MF, Cohen RA, Faller DV. A physiological delay in human fetal hemoglobin switching is associated with specific globin DNA hypomethylation. FEBS Lett 1988; 228:139–43 [DOI] [PubMed] [Google Scholar]
- 28.Busslinger M, Hurst J, Flavell RA. DNA methylation and the regulation of globin gene expression. Cell 1983; 34:197–206 [DOI] [PubMed] [Google Scholar]
- 29.Comi P, Ottolenghi S, Giglioni B, Migliaccio G, Migliaccio AR, Bassano E, Amadori S, Mastroberardino G, Peschle C. Bromodeoxyuridine treatment of normal adult erythroid colonies: an in vitro model for reactivation of human fetal globin genes. Blood 1986; 68:1036–41 [PubMed] [Google Scholar]
- 30.Lavelle D, Saunthararajah Y, Vaitkus K, Singh M, Banzon V, Phiasivongsva P, Redkar S, Kanekal S, Bearss D, Shi C, Inloes R, DeSimone J. S110, a novel decitabine dinucleotide, increases fetal hemoglobin levels in baboons (P. anubis). J Transl Med 2010; 8:92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Molokie R, Lavelle D, Gowhari M, Pacini M, Krauz L, Hassan J, Ibanez V, Ruiz MA, Ng KP, Woost P, Radivoyevitch T, Pacelli D, Fada S, Rump M, Hsieh M, Tisdale JF, Jacobberger J, Phelps M, Engel JD, Saraf S, Hsu LL, Gordeuk V, DeSimone J, Saunthararajah Y. Oral tetrahydrouridine and decitabine for non-cytotoxic epigenetic gene regulation in sickle cell disease: a randomized phase 1 study. PLoS Med 2017; 14:e1002382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Singal R, Wang SZ, Sargent T, Zhu SZ, Ginder GD. Methylation of promoter proximal-transcribed sequences of an embryonic globin gene inhibits transcription in primary erythroid cells and promotes formation of a cell type-specific methyl cytosine binding complex. J Biol Chem 2002; 277:1897–905 [DOI] [PubMed] [Google Scholar]
- 33.Ginder GD, Gnanapragasam MN, Mian OY. The role of the epigenetic signal, DNA methylation, in gene regulation during erythroid development. Curr Top Dev Biol 2008; 82:85–116 [DOI] [PubMed] [Google Scholar]
- 34.Rivers A, Vaitkus K, Ibanez V, Ruiz MA, Jagadeeswaran R, Saunthararajah Y, Cui S, Engel JD, DeSimone J, Lavelle D. The LSD1 inhibitor RN-1 recapitulates the fetal pattern of hemoglobin synthesis in baboons (P. anubis). Haematologica 2016; 101:688–97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Forrester WC, Thompson C, Elder JT, Groudine M. A developmentally stable chromatin structure in the human beta-globin gene cluster. Proc Natl Acad Sci U S A 1986; 83:1359–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Goren A, Simchen G, Fibach E, Szabo PE, Tanimoto K, Chakalova L, Pfeifer GP, Fraser PJ, Engel JD, Cedar H. Fine tuning of globin gene expression by DNA methylation. PLoS One 2006; 1:e46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Singh M, Lavelle D, Vaitkus K, Mahmud N, Hankewych M, DeSimone J. The gamma-globin gene promoter progressively demethylates as the hematopoietic stem progenitor cells differentiate along the erythroid lineage in baboon fetal liver and adult bone marrow. Exp Hematol 2007; 35:48–55 [DOI] [PubMed] [Google Scholar]
- 38.Suzuki M, Yamamoto M, Engel JD. Fetal globin gene repressors as drug targets for molecular therapies to treat the beta-globinopathies. Mol Cell Biol 2014; 34:3560–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhao Q, Rank G, Tan YT, Li H, Moritz RL, Simpson RJ, Cerruti L, Curtis DJ, Patel DJ, Allis CD, Cunningham JM, Jane SM. PRMT5-mediated methylation of histone H4R3 recruits DNMT3A, coupling histone and DNA methylation in gene silencing. Nat Struct Mol Biol 2009; 16:304–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rank G, Cerruti L, Simpson RJ, Moritz RL, Jane SM, Zhao Q. Identification of a PRMT5-dependent repressor complex linked to silencing of human fetal globin gene expression. Blood 2010; 116:1585–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Banzon V, Ibanez V, Vaitkus K, Ruiz MA, Peterson K, DeSimone J, Lavelle D. siDNMT1 increases gamma-globin expression in chemical inducer of dimerization (CID)-dependent mouse betaYAC bone marrow cells and in baboon erythroid progenitor cell cultures. Exp Hematol 2011; 39:26–36.e1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Galarneau G, Palmer CD, Sankaran VG, Orkin SH, Hirschhorn JN, Lettre G. Fine-mapping at three loci known to affect fetal hemoglobin levels explains additional genetic variation. Nat Genet 2010; 42:1049–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Thein SL, Menzel S, Peng X, Best S, Jiang J, Close J, Silver N, Gerovasilli A, Ping C, Yamaguchi M, Wahlberg K, Ulug P, Spector TD, Garner C, Matsuda F, Farrall M, Lathrop M. Intergenic variants of HBS1L-MYB are responsible for a major quantitative trait locus on chromosome 6q23 influencing fetal hemoglobin levels in adults. Proc Natl Acad Sci U S A 2007; 104:11346–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Menzel S, Garner C, Gut I, Matsuda F, Yamaguchi M, Heath S, Foglio M, Zelenika D, Boland A, Rooks H, Best S, Spector TD, Farrall M, Lathrop M, Thein SL. A QTL influencing F cell production maps to a gene encoding a zinc-finger protein on chromosome 2p15. Nat Genet 2007; 39:1197–9 [DOI] [PubMed] [Google Scholar]
- 45.Uda M, Galanello R, Sanna S, Lettre G, Sankaran VG, Chen W, Usala G, Busonero F, Maschio A, Albai G, Piras MG, Sestu N, Lai S, Dei M, Mulas A, Crisponi L, Naitza S, Asunis I, Deiana M, Nagaraja R, Perseu L, Satta S, Cipollina MD, Sollaino C, Moi P, Hirschhorn JN, Orkin SH, Abecasis GR, Schlessinger D, Cao A. Genome-wide association study shows BCL11A associated with persistent fetal hemoglobin and amelioration of the phenotype of beta-thalassemia. Proc Natl Acad Sci U S A 2008; 105:1620–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lettre G, Sankaran VG, Bezerra MA, Araujo AS, Uda M, Sanna S, Cao A, Schlessinger D, Costa FF, Hirschhorn JN, Orkin SH. DNA polymorphisms at the BCL11A, HBS1L-MYB, and beta-globin loci associate with fetal hemoglobin levels and pain crises in sickle cell disease. Proc Natl Acad Sci U S A 2008; 105:11869–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bauer DE, Kamran SC, Orkin SH. Reawakening fetal hemoglobin: prospects for new therapies for the beta-globin disorders. Blood 2012; 120:2945–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sankaran VG, Menne TF, Xu J, Akie TE, Lettre G, Van Handel B, Mikkola HK, Hirschhorn JN, Cantor AB, Orkin SH. Human fetal hemoglobin expression is regulated by the developmental stage-specific repressor BCL11A. Science 2008; 322:1839–42 [DOI] [PubMed] [Google Scholar]
- 49.Makani J, Menzel S, Nkya S, Cox SE, Drasar E, Soka D, Komba AN, Mgaya J, Rooks H, Vasavda N, Fegan G, Newton CR, Farrall M, Thein SL. Genetics of fetal hemoglobin in Tanzanian and British patients with sickle cell anemia. Blood 2011; 117:1390–2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bieker JJ. Probing the onset and regulation of erythroid cell-specific gene expression. Mt Sinai J Med 2005; 72:333–8 [PubMed] [Google Scholar]
- 51.Zhou D, Liu K, Sun CW, Pawlik KM, Townes TM. KLF1 regulates BCL11A expression and gamma- to beta-globin gene switching. Nat Genet 2010; 42:742–4 [DOI] [PubMed] [Google Scholar]
- 52.Miller IJ, Bieker JJ. A novel, erythroid cell-specific murine transcription factor that binds to the CACCC element and is related to the kruppel family of nuclear proteins. Mol Cell Biol 1993; 13:2776–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shariati L, Khanahmad H, Salehi M, Hejazi Z, Rahimmanesh I, Tabatabaiefar MA, Modarressi MH. Genetic disruption of the KLF1 gene to overexpress the gamma-globin gene using the CRISPR/Cas9 system. J Gene Med 2016; 18:294–301 [DOI] [PubMed] [Google Scholar]
- 54.Tallack MR, Perkins AC. Three fingers on the switch: Kruppel-like factor 1 regulation of gamma-globin to beta-globin gene switching. Curr Opin Hematol 2013; 20:193–200 [DOI] [PubMed] [Google Scholar]
- 55.Borg J, Papadopoulos P, Georgitsi M, Gutierrez L, Grech G, Fanis P, Phylactides M, Verkerk AJ, van der Spek PJ, Scerri CA, Cassar W, Galdies R, van Ijcken W, Ozgur Z, Gillemans N, Hou J, Bugeja M, Grosveld FG, von Lindern M, Felice AE, Patrinos GP, Philipsen S. Haploinsufficiency for the erythroid transcription factor KLF1 causes hereditary persistence of fetal hemoglobin. Nat Genet 2010; 42:801–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tallack MR, Magor GW, Dartigues B, Sun L, Huang S, Fittock JM, Fry SV, Glazov EA, Bailey TL, Perkins AC. Novel roles for KLF1 in erythropoiesis revealed by mRNA-seq. Genome Res 2012; 22:2385–98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Siatecka M, Bieker JJ. The multifunctional role of EKLF/KLF1 during erythropoiesis. Blood 2011; 118:2044–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Jiang J, Best S, Menzel S, Silver N, Lai MI, Surdulescu GL, Spector TD, Thein SL. cMYB is involved in the regulation of fetal hemoglobin production in adults. Blood 2006; 108:1077–83 [DOI] [PubMed] [Google Scholar]
- 59.Bianchi E, Zini R, Salati S, Tenedini E, Norfo R, Tagliafico E, Manfredini R, Ferrari S. c-myb supports erythropoiesis through the transactivation of KLF1 and LMO2 expression. Blood 2010; 116:e99–110. [DOI] [PubMed] [Google Scholar]
- 60.Sankaran VG, Menne TF, Scepanovic D, Vergilio JA, Ji P, Kim J, Thiru P, Orkin SH, Lander ES, Lodish HF. MicroRNA-15a and -16-1 act via MYB to elevate fetal hemoglobin expression in human trisomy 13. Proc Natl Acad Sci U S A 2011; 108:1519–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Masuda T, Wang X, Maeda M, Canver MC, Sher F, Funnell AP, Fisher C, Suciu M, Martyn GE, Norton LJ, Zhu C, Kurita R, Nakamura Y, Xu J, Higgs DR, Crossley M, Bauer DE, Orkin SH, Kharchenko PV, Maeda T. Transcription factors LRF and BCL11A independently repress expression of fetal hemoglobin. Science 2016; 351:285–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wu X, Zhang Y. TET-mediated active DNA demethylation: mechanism, function and beyond. Nat Rev Genet 2017; 18:517–34 [DOI] [PubMed] [Google Scholar]
- 63.Sengupta PK, Lavelle D, DeSimone J. Increased binding of Sp1 to the gamma-globin gene promoter upon site-specific cytosine methylation. Am J Hematol 1994; 46:169–72 [DOI] [PubMed] [Google Scholar]
- 64.Jane SM, Gumucio DL, Ney PA, Cunningham JM, Nienhuis AW. Methylation-enhanced binding of Sp1 to the stage selector element of the human gamma-globin gene promoter may regulate development specificity of expression. Mol Cell Biol 1993; 13:3272–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Labbe RM, Holowatyj A, Yang ZQ. Histone lysine demethylase (KDM) subfamily 4: structures, functions and therapeutic potential. Am J Transl Res 2013; 6:1–15 [PMC free article] [PubMed] [Google Scholar]
- 66.Xu J, Bauer DE, Kerenyi MA, Vo TD, Hou S, Hsu YJ, Yao H, Trowbridge JJ, Mandel G, Orkin SH. Corepressor-dependent silencing of fetal hemoglobin expression by BCL11A. Proc Natl Acad Sci U S A 2013; 110:6518–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Shi L, Cui S, Engel JD, Tanabe O. Lysine-specific demethylase 1 is a therapeutic target for fetal hemoglobin induction. Nat Med 2013; 19:291–4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Rivers A, Vaitkus K, Jagadeeswaran R, Ruiz MA, Ibanez V, Ciceri F, Cavalcanti F, Molokie RE, Saunthararajah Y, Engel JD, DeSimone J, Lavelle D. Oral administration of the LSD1 inhibitor ORY-3001 increases fetal hemoglobin in sickle cell mice and baboons. Exp Hematol 2018; 67:60–4.e2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Du Q, Luu PL, Stirzaker C, Clark SJ. Methyl-CpG-binding domain proteins: readers of the epigenome. Epigenomics 2015; 7:1051–73 [DOI] [PubMed] [Google Scholar]
- 70.Rivers A, Molokie R, Lavelle D. A new target for fetal hemoglobin reactivation. Haematologica 2019; 104:2325–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Rupon JW, Wang SZ, Gaensler K, Lloyd J, Ginder GD. Methyl binding domain protein 2 mediates gamma-globin gene silencing in adult human betaYAC transgenic mice. Proc Natl Acad Sci U S A 2006; 103:6617–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Yu X, Azzo A, Bilinovich SM, Li X, Dozmorov M, Kurita R, Nakamura Y, Williams DC, Jr., Ginder GD. Disruption of the MBD2-NuRD complex but not MBD3-NuRD induces high level HbF expression in human adult erythroid cells. Haematologica 2019; 104:2361–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Yu L, Jearawiriyapaisarn N, Lee MP, Hosoya T, Wu Q, Myers G, Lim KC, Kurita R, Nakamura Y, Vojtek AB, Rual JF, Engel JD. BAP1 regulation of the key adaptor protein NCoR1 is critical for gamma-globin gene repression. Genes Dev 2018; 32:1537–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Martin H, Steinberg DHKC, Dover GJ, Sebastiani P, Alsultan A. Fetal hemoglobin in sickle cell anemia: a glass half full? Blood 2014; 123:481–5 [DOI] [PubMed] [Google Scholar]
- 75.Koshy M, Dorn L, Bressler L, Molokie R, Lavelle D, Talischy N, Hoffman R, van Overveld W, DeSimone J. 2-deoxy 5-azacytidine and fetal hemoglobin induction in sickle cell anemia. Blood 2000; 96:2379–84 [PubMed] [Google Scholar]
- 76.Stomper J, Ihorst G, Suciu S, Sander PN, Becker H, Wijermans PW, Plass C, Weichenhan D, Bisse E, Claus R, Lubbert M. Fetal hemoglobin induction during decitabine treatment of elderly patients with high-risk myelodysplastic syndrome or acute myeloid leukemia: a potential dynamic biomarker of outcome. Haematologica 2019; 104:59–69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Mabaera R, Greene MR, Richardson CA, Conine SJ, Kozul CD, Lowrey CH. Neither DNA hypomethylation nor changes in the kinetics of erythroid differentiation explain 5-azacytidine's ability to induce human fetal hemoglobin. Blood 2008; 111:411–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Carr BI, Rahbar S, Doroshow JH, Blayney D, Goldberg D, Leong L, Asmeron Y. Fetal hemoglobin gene activation in a phase II study of 5,6-dihydro-5-azacytidine for bronchogenic carcinoma. Cancer Res 1987; 47:4199–201 [PubMed] [Google Scholar]
- 79.Press KR, Uy N, Keefer J, Gore SD, Carraway HE, Sakoian S, Prebet T. Clinical evaluation of combined azacitidine and entinostat on the induction of fetal hemoglobin in patients with acute myeloid leukemias and myelodysplastic syndromes. Leuk Lymphoma 2018; 59:755–7 [DOI] [PubMed] [Google Scholar]
- 80.Kalantri SA, Ray R, Chattopadhyay A, Bhattacharjee S, Biswas A, Bhattacharyya M. Efficacy of decitabine as hemoglobin F inducer in HbE/beta-thalassemia. Ann Hematol 2018; 97:1689–94 [DOI] [PubMed] [Google Scholar]
- 81.Gambari R, del Senno L, Barbieri R, Viola L, Tripodi M, Raschella G, Fantoni A. Human leukemia K-562 cells: induction of erythroid differentiation by 5-azacytidine. Cell Differ 1984; 14:87–97 [DOI] [PubMed] [Google Scholar]
- 82.Charache S, Dover G, Smith K, Talbot CC, Jr., Moyer M, Boyer S. Treatment of sickle cell anemia with 5-azacytidine results in increased fetal hemoglobin production and is associated with nonrandom hypomethylation of DNA around the gamma-Delta-beta-globin gene complex. Proc Natl Acad Sci U S A 1983; 80:4842–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.DeSimone J, Heller P, Schimenti JC, Duncan CH. Fetal hemoglobin production in adult baboons by 5-azacytidine or by phenylhydrazine-induced hemolysis is associated with hypomethylation of globin gene DNA. Prog Clin Biol Res 1983; 134:489–500 [PubMed] [Google Scholar]
- 84.DeSimone J, Heller P, Hall L, Zwiers D. 5-Azacytidine stimulates fetal hemoglobin synthesis in anemic baboons. Proc Natl Acad Sci U S A 1982; 79:4428–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Christman JK. 5-Azacytidine and 5-aza-2'-deoxycytidine as inhibitors of DNA methylation: mechanistic studies and their implications for cancer therapy. Oncogene 2002; 21:5483–95 [DOI] [PubMed] [Google Scholar]
- 86.Ley TJ, DeSimone J, Anagnou NP, Keller GH, Humphries RK, Turner PH, Young NS, Keller P, Nienhuis AW. 5-azacytidine selectively increases gamma-globin synthesis in a patient with beta+ thalassemia. N Engl J Med 1982; 307:1469–75 [DOI] [PubMed] [Google Scholar]
- 87.Ley TJ, DeSimone J, Noguchi CT, Turner PH, Schechter AN, Heller P, Nienhuis AW. 5-Azacytidine increases gamma-globin synthesis and reduces the proportion of dense cells in patients with sickle cell anemia. Blood 1983; 62:370–80 [PubMed] [Google Scholar]
- 88.Akpan I, Banzon V, Ibanez V, Vaitkus K, DeSimone J, Lavelle D. Decitabine increases fetal hemoglobin in papio anubis by increasing gamma-globin gene transcription. Exp Hematol 2010; 38:989–93.e1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Dover GJ, Charache SH, Boyer SH, Talbot CC, Jr., Smith KD. 5-Azacytidine increases fetal hemoglobin production in a patient with sickle cell disease. Prog Clin Biol Res 1983; 134:475–88 [PubMed] [Google Scholar]
- 90.Ley TJ, Anagnou NP, Noguchi CT, Schechter AN, DeSimone J, Heller P, Nienhuis AW. DNA methylation and globin gene expression in patients treated with 5-azacytidine. Prog Clin Biol Res 1983; 134:457–74 [PubMed] [Google Scholar]
- 91.Chondrou V, Stavrou EF, Markopoulos G, Kouraklis-Symeonidis A, Fotopoulos V, Symeonidis A, Vlachaki E, Chalkia P, Patrinos GP, Papachatzopoulou A, Sgourou A. Impact of ZBTB7A hypomethylation and expression patterns on treatment response to hydroxyurea. Hum Genom 2018; 12:45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Camiener GW, Smith CG. Studies of the enzymatic deamination of cytosine arabinoside. I. Enzyme distribution and species specificity. Biochem Pharmacol 1965; 14:1405–16 [DOI] [PubMed] [Google Scholar]
- 93.Ebrahem Q, Mahfouz RZ, Ng KP, Saunthararajah Y. High cytidine deaminase expression in the liver provides sanctuary for cancer cells from decitabine treatment effects. Oncotarget 2012; 3:1137–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Gao C, Wang J, Li Y, Zhao H, Li R, Hou L, Zhang Y, Tian S, Liang H, Wang C, Chen X, Wang J. Incidence and risk of hematologic toxicities with hypomethylating agents in the treatment of myelodysplastic syndromes and acute myeloid leukopenia: a systematic review and meta-analysis. Medicine 2018; 97:e11860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Atweh GF, DeSimone J, Saunthararajah Y, Fathallah H, Weinberg RS, Nagel RL, Fabry ME, Adams RJ. Hemoglobinopathies. Hematology Am Soc Hematol Educ Program 2003;14–39. DOI: 10.1182/asheducation-2003.1.14 [DOI] [PubMed] [Google Scholar]
- 96.Zauri M, Berridge G, Thezenas ML, Pugh KM, Goldin R, Kessler BM, Kriaucionis S. CDA directs metabolism of epigenetic nucleosides revealing a therapeutic window in cancer. Nature 2015; 524:114–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.https://www.fda.gov/drugs/drug-approvals-and-databases/fda-approves-oral-combination-decitabine-and-cedazuridine-myelodysplastic-syndromes (2020, accessed 15 June 2021)
- 98.Lavelle D, Vaitkus K, Ling Y, Ruiz MA, Mahfouz R, Ng KP, Negrotto S, Smith N, Terse P, Engelke KJ, Covey J, Chan KK, Desimone J, Saunthararajah Y. Effects of tetrahydrouridine on pharmacokinetics and pharmacodynamics of oral decitabine. Blood 2012; 119:1240–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Wu H, Zhang Y. Reversing DNA methylation: mechanisms, genomics, and biological functions. Cell 2014; 156:45–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.He Y, Rank G, Zhang M, Ju J, Liu R, Xu Z, Brown F, Cerruti L, Ma C, Tan R, Jane SM, Zhao Q. Induction of human fetal hemoglobin expression by adenosine-2',3'-dialdehyde. J Transl Med 2013; 11:14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Gilmartin AG, Groy A, Gore ER, Atkins C, Long ER, 3rd, Montoute MN, Wu Z, Halsey W, McNulty DE, Ennulat D, Rueda L, Pappalardi M, Kruger RG, McCabe MT, Raoof A, Butlin R, Stowell A, Cockerill M, Waddell I, Ogilvie D, Luengo J, Jordan A, Benowitz AB. In vitro and in vivo induction of fetal hemoglobin with a reversible and selective DNMT1 inhibitor. Haematologica 2020. DOI: 10.3324/haematol.2020.248658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Miller JL, Grant PA. The role of DNA methylation and histone modifications in transcriptional regulation in humans. Subcell Biochem 2013; 61:289–317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K. High-resolution profiling of histone methylations in the human genome. Cell 2007; 129:823–37 [DOI] [PubMed] [Google Scholar]
- 104.Klose RJ, Yan Q, Tothova Z, Yamane K, Erdjument-Bromage H, Tempst P, Gilliland DG, Zhang Y, Kaelin WG., Jr., The retinoblastoma binding protein RBP2 is an H3K4 demethylase. Cell 2007; 128:889–900 [DOI] [PubMed] [Google Scholar]
- 105.Ginder GD, Whitters MJ, Pohlman JK, Burns LJ. Transcriptional activation of an embryonic globin gene in adult erythroid cells in vivo. Prog Clin Biol Res 1985; 191:463–74 [PubMed] [Google Scholar]
- 106.Perrine SP, Rudolph A, Faller DV, Roman C, Cohen RA, Chen SJ, Kan YW. Butyrate infusions in the ovine fetus delay the biologic clock for globin gene switching. Proc Natl Acad Sci U S A 1988; 85:8540–2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Forsberg EC, Downs KM, Christensen HM, Im H, Nuzzi PA, Bresnick EH. Developmentally dynamic histone acetylation pattern of a tissue-specific chromatin domain. Proc Natl Acad Sci U S A 2000; 97:14494–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Perrine SP, Swerdlow P, Faller DV, Qin G, Rudolph AM, Reczek J, Kan YW. Butyric acid modulates developmental globin gene switching in man and sheep. Adv Exp Med Biol 1989; 271:177–83 [DOI] [PubMed] [Google Scholar]
- 109.Aerbajinai W, Zhu J, Gao Z, Chin K, Rodgers GP. Thalidomide induces gamma-globin gene expression through increased reactive oxygen species-mediated p38 MAPK signaling and histone H4 acetylation in adult erythropoiesis. Blood 2007; 110:2864–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Zhao Z, Shilatifard A. Epigenetic modifications of histones in cancer. Genome Biol 2019; 20:245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Portela A, Esteller M. Epigenetic modifications and human disease. Nat Biotechnol 2010; 28:1057–68 [DOI] [PubMed] [Google Scholar]
- 112.Dai Y, Chen T, Ijaz H, Cho EH, Steinberg MH. SIRT1 activates the expression of fetal hemoglobin genes. Am J Hematol 2017; 92:1177–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Woon Kim Y, Kim S, Geun Kim C, Kim A. The distinctive roles of erythroid specific activator GATA-1 and NF-E2 in transcription of the human fetal gamma-globin genes. Nucleic Acids Res 2011; 39:6944–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Layon ME, Ackley CJ, West RJ, Lowrey CH. Expression of GATA-1 in a non-hematopoietic cell line induces beta-globin locus control region chromatin structure remodeling and an erythroid pattern of gene expression. J Mol Biol 2007; 366:737–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Constantoulakis P, Knitter G, Stamatoyannopoulos G. On the induction of fetal hemoglobin by butyrates: in vivo and in vitro studies with sodium butyrate and comparison of combination treatments with 5-AzaC and AraC. Blood 1989; 74:1963–71 [PubMed] [Google Scholar]
- 116.Perrine SP, Ginder GD, Faller DV, Dover GH, Ikuta T, Witkowska HE, Cai SP, Vichinsky EP, Olivieri NF. A short-term trial of butyrate to stimulate fetal-globin-gene expression in the beta-globin disorders. N Engl J Med 1993; 328:81–6 [DOI] [PubMed] [Google Scholar]
- 117.Atweh GF, Sutton M, Nassif I, Boosalis V, Dover GJ, Wallenstein S, Wright E, McMahon L, Stamatoyannopoulos G, Faller DV, Perrine SP. Sustained induction of fetal hemoglobin by pulse butyrate therapy in sickle cell disease. Blood 1999; 93:1790–7 [PMC free article] [PubMed] [Google Scholar]
- 118.Bradner JE, Mak R, Tanguturi SK, Mazitschek R, Haggarty SJ, Ross K, Chang CY, Bosco J, West N, Morse E, Lin K, Shen JP, Kwiatkowski NP, Gheldof N, Dekker J, DeAngelo DJ, Carr SA, Schreiber SL, Golub TR, Ebert BL. Chemical genetic strategy identifies histone deacetylase 1 (HDAC1) and HDAC2 as therapeutic targets in sickle cell disease. Proc Natl Acad Sci U S A 2010; 107:12617–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Pace BS, Qian XH, Sangerman J, Ofori-Acquah SF, Baliga BS, Han J, Critz SD. p38 MAP kinase activation mediates gamma-globin gene induction in erythroid progenitors. Exp Hematol 2003; 31:1089–96 [DOI] [PubMed] [Google Scholar]
- 120.Hsiao CH, Li W, Lou TF, Baliga BS, Pace BS. Fetal hemoglobin induction by histone deacetylase inhibitors involves generation of reactive oxygen species. Exp Hematol 2006; 34:264–73 [DOI] [PubMed] [Google Scholar]
- 121.Witt O, Monkemeyer S, Ronndahl G, Erdlenbruch B, Reinhardt D, Kanbach K, Pekrun A. Induction of fetal hemoglobin expression by the histone deacetylase inhibitor apicidin. Blood 2003; 101:2001–7 [DOI] [PubMed] [Google Scholar]
- 122.Debrand E, Chakalova L, Miles J, Dai YF, Goyenechea B, Dye S, Osborne CS, Horton A, Harju-Baker S, Pink RC, Caley D, Carter DRF, Peterson KR, Fraser P. An intergenic non-coding RNA promoter required for histone modifications in the human beta-globin chromatin domain. PLoS One 2019; 14:e0217532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Qian X, Chen J, Zhao D, Guo L, Qian X. Plastrum testudinis induces gamma-globin gene expression through epigenetic histone modifications within the gamma-globin gene promoter via activation of the p38 MAPK signaling pathway. Int J Mol Med 2013; 31:1418–28 [DOI] [PubMed] [Google Scholar]
- 124.Chang KH, Fang X, Wang H, Huang A, Cao H, Yang Y, Bonig H, Stamatoyannopoulos JA, Papayannopoulou T. Epigenetic modifications and chromosome conformations of the beta globin locus throughout development. Stem Cell Rev Rep 2013; 9:397–407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.van Dijk TB, Gillemans N, Pourfarzad F, van Lom K, von Lindern M, Grosveld F, Philipsen S. Fetal globin expression is regulated by friend of Prmt1. Blood 2010; 116:4349–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Li X, Hu X, Patel B, Zhou Z, Liang S, Ybarra R, Qiu Y, Felsenfeld G, Bungert J, Huang S. H4R3 methylation facilitates beta-globin transcription by regulating histone acetyltransferase binding and H3 acetylation. Blood 2010; 115:2028–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Ju J, Wang Y, Liu R, Zhang Y, Xu Z, Wang Y, Wu Y, Liu M, Cerruti L, Zou F, Ma C, Fang M, Tan R, Jane SM, Zhao Q. Human fetal globin gene expression is regulated by LYAR. Nucleic Acids Res 2014; 42:9740–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Perrine SP, Olivieri NF, Faller DV, Vichinsky EP, Dover GJ, Ginder GD. Butyrate derivatives. New agents for stimulating fetal globin production in the beta-globin disorders. Am J Pediatr Hematol Oncol 1994; 16:67–71 [PubMed] [Google Scholar]
- 129.Weinberg RS, Ji X, Sutton M, Perrine S, Galperin Y, Li Q, Liebhaber SA, Stamatoyannopoulos G, Atweh GF. Butyrate increases the efficiency of translation of gamma-globin mRNA. Blood 2005; 105:1807–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Cao H. Pharmacological induction of fetal hemoglobin synthesis using histone deacetylase inhibitors. Hematology 2004; 9:223–33 [DOI] [PubMed] [Google Scholar]
- 131.Sher GD, Ginder GD, Little J, Yang S, Dover GJ, Olivieri NF. Extended therapy with intravenous arginine butyrate in patients with beta-hemoglobinopathies. N Engl J Med 1995; 332:1606–10 [DOI] [PubMed] [Google Scholar]
- 132.Hudgins WR, Fibach E, Safaya S, Rieder RF, Miller AC, Samid D. Transcriptional upregulation of gamma-globin by phenylbutyrate and analogous aromatic fatty acids. Biochem Pharmacol 1996; 52:1227–33 [DOI] [PubMed] [Google Scholar]
- 133.Fucharoen S, Inati A, Siritanaratku N, Thein SL, Wargin WC, Koussa S, Taher A, Chaneim N, Boosalis M, Berenson R, Perrine SP. A randomized phase I/II trial of HQK-1001, an oral fetal globin gene inducer, in beta-thalassaemia intermedia and HbE/beta-thalassaemia. Br J Haematol 2013; 161:587–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Saki N, Abroun S, Hajizamani S, Rahim F, Shahjahani M. Association of chromosomal translocation and MiRNA expression with the pathogenesis of multiple myeloma. Cell J 2014; 16:99–110 [PMC free article] [PubMed] [Google Scholar]
- 135.Kim VN. Small RNAs: classification, biogenesis, and function. Mol Cells 2005; 19:1–15 [PubMed] [Google Scholar]
- 136.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 2004; 116:281–97 [DOI] [PubMed] [Google Scholar]
- 137.Bentwich I, Avniel A, Karov Y, Aharonov R, Gilad S, Barad O, Barzilai A, Einat P, Einav U, Meiri E, Sharon E, Spector Y, Bentwich Z. Identification of hundreds of conserved and nonconserved human microRNAs. Nat Genet 2005; 37:766–70 [DOI] [PubMed] [Google Scholar]
- 138.Li Y, Liu D, Zhang X, Li Z, Ye Y, Liu Q, Shen J, Chen Z, Huang H, Liang Y, Han X, Liu J, An X, Mohandas N, Xu X. miR-326 regulates HbF synthesis by targeting EKLF in human erythroid cells. Exp Hematol 2018; 63:33–40.e2 [DOI] [PubMed] [Google Scholar]
- 139.Alijani S, Alizadeh S, Kazemi A, Khatib ZK, Soleimani M, Rezvani M, Minayi N, Karami F, Tayebi B. Evaluation of the effect of miR-26b up-Regulation on HbF expression in erythroleukemic K-562 cell line. Avicenna J Med Biotechnol 2014; 6:53–6 [PMC free article] [PubMed] [Google Scholar]
- 140.Pule GD, Mowla S, Novitzky N, Wonkam A. Hydroxyurea down-regulates BCL11A, KLF-1 and MYB through miRNA-mediated actions to induce gamma-globin expression: implications for new therapeutic approaches of sickle cell disease. Clin Transl Med 2016; 5:15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Lulli V, Romania P, Morsilli O, Cianciulli P, Gabbianelli M, Testa U, Giuliani A, Marziali G. MicroRNA-486-3p regulates gamma-globin expression in human erythroid cells by directly modulating BCL11A. PLoS One 2013; 8:e60436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Guda S, Brendel C, Renella R, Du P, Bauer DE, Canver MC, Grenier JK, Grimson AW, Kamran SC, Thornton J, de Boer H, Root DE, Milsom MD, Orkin SH, Gregory RI, Williams DA. miRNA-embedded shRNAs for lineage-specific BCL11A knockdown and hemoglobin F induction. Mol Ther 2015; 23:1465–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Yuan JY, Wang F, Yu J, Yang GH, Liu XL, Zhang JW. MicroRNA-223 reversibly regulates erythroid and megakaryocytic differentiation of K562 cells. J Cell Mol Med 2009; 13:4551–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Bianchi N, Zuccato C, Lampronti I, Borgatti M, Gambari R. Expression of miR-210 during erythroid differentiation and induction of gamma-globin gene expression. BMB Rep 2009; 42:493–9 [DOI] [PubMed] [Google Scholar]
- 145.Gasparello J, Fabbri E, Bianchi N, Breveglieri G, Zuccato C, Borgatti M, Gambari R, Finotti A. BCL11A mRNA targeting by miR-210: a possible network regulating gamma-globin gene expression. Int J Mol Sci 2017; 18. DOI: 10.3390/ijms18122530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Bianchi N, Finotti A, Ferracin M, Lampronti I, Zuccato C, Breveglieri G, Brognara E, Fabbri E, Borgatti M, Negrini M, Gambari R. Increase of microRNA-210, decrease of raptor gene expression and alteration of mammalian target of rapamycin regulated proteins following mithramycin treatment of human erythroid cells. PLoS One 2015; 10:e0121567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Ward CM, Li B, Pace BS. Original research: stable expression of miR-34a mediates fetal hemoglobin induction in K562 cells. Exp Biol Med 2016; 241:719–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Ma Y, Wang B, Jiang F, Wang D, Liu H, Yan Y, Dong H, Wang F, Gong B, Zhu Y, Dong L, Yin H, Zhang Z, Zhao H, Wu Z, Zhang J, Zhou J, Yu J. A feedback loop consisting of microRNA 23a/27a and the beta-like globin suppressors KLF3 and SP1 regulates globin gene expression. Mol Cell Biol 2013; 33:3994–4007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Walker AL, Steward S, Howard TA, Mortier N, Smeltzer M, Wang YD, Ware RE. Epigenetic and molecular profiles of erythroid cells after hydroxyurea treatment in sickle cell anemia. Blood 2011; 118:5664–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Zhu X, Li B, Pace BS. NRF2 mediates gamma-globin gene regulation and fetal hemoglobin induction in human erythroid progenitors. Haematologica 2017; 102:e285–e88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Lee YT, de Vasconcellos JF, Yuan J, Byrnes C, Noh SJ, Meier ER, Kim KS, Rabel A, Kaushal M, Muljo SA, Miller JL. LIN28B-mediated expression of fetal hemoglobin and production of fetal-like erythrocytes from adult human erythroblasts ex vivo. Blood 2013; 122:1034–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Kollinerova S, Vassanelli S, Modriansky M. The role of miR-29 family members in malignant hematopoiesis. Biomed Pap Med Fac Univ Palacky Olomouc Czech Repub 2014; 158:489–501 [DOI] [PubMed] [Google Scholar]
- 153.Fiserova B, Kubiczkova L, Sedlarikova L, Hajek R, Sevcikova S. The miR-29 family in hematological malignancies. Biomed Pap Med Fac Univ Palacky Olomouc Czech Repub 2015; 159:184–91 [DOI] [PubMed] [Google Scholar]
- 154.Chen Y, Gao DY, Huang L. In vivo delivery of miRNAs for cancer therapy: challenges and strategies. Adv Drug Deliv Rev 2015; 81:128–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Osborn MF, Khvorova A. Improving siRNA delivery in vivo through lipid conjugation. Nucleic Acid Ther 2018; 28:128–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Gaibelet G, Allart S, Terce F, Azalbert V, Bertrand-Michel J, Hamdi S, Collet X, Orlowski S. Specific cellular incorporation of a pyrene-labelled cholesterol: lipoprotein-mediated delivery toward ordered intracellular membranes. PLoS One 2015; 10:e0121563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Byrne M, Tzekov R, Wang Y, Rodgers A, Cardia J, Ford G, Holton K, Pandarinathan L, Lapierre J, Stanney W, Bulock K, Shaw S, Libertine L, Fettes K, Khvorova A, Kaushal S, Pavco P. Novel hydrophobically modified asymmetric RNAi compounds (sd-rxRNA) demonstrate robust efficacy in the eye. J Ocul Pharmacol Ther 2013; 29:855–64 [DOI] [PubMed] [Google Scholar]
- 158.Rupaimoole R, Slack FJ. MicroRNA therapeutics: towards a new era for the management of cancer and other diseases. Nat Rev Drug Discov 2017; 16:203–22 [DOI] [PubMed] [Google Scholar]
- 159.Lopez-Gonzalez MJ, Landry M, Favereaux A. MicroRNA and chronic pain: from mechanisms to therapeutic potential. Pharmacol Ther 2017; 180:1–15 [DOI] [PubMed] [Google Scholar]
- 160.McClure JJ, Li X, Chou CJ. Advances and challenges of HDAC inhibitors in cancer therapeutics. Adv Cancer Res 2018; 138:183–211 [DOI] [PubMed] [Google Scholar]
- 161.Issa JP, Kantarjian HM. Targeting DNA methylation. Clin Cancer Res 2009; 15:3938–46 [DOI] [PMC free article] [PubMed] [Google Scholar]