Fig. 9.
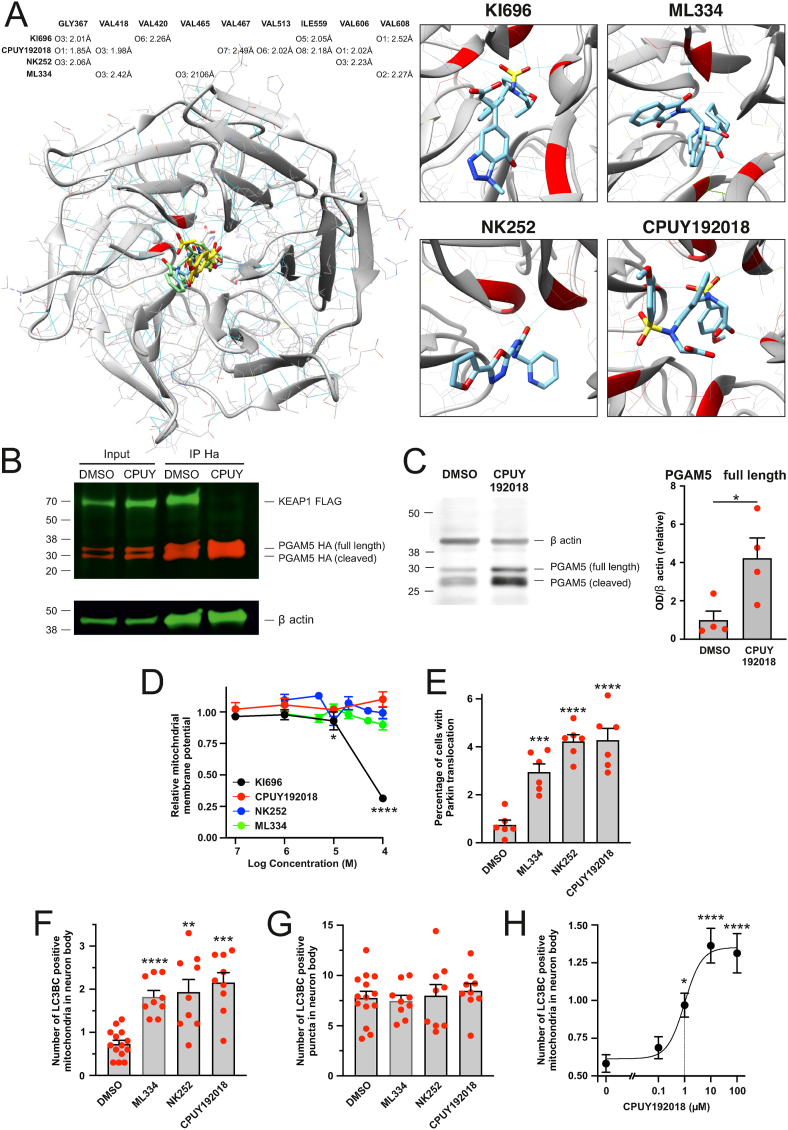
Inhibitors of KEAP1–PGAM5 protein-protein interaction enhance mitophagy. A. Molecular docking simulation showing energetically most favorable interactions between a KEAP1 and non-electrophilic KEAP1 inhibitors. The table is showing hydrogen-bond pairings in KEAP1-inhibitor complexes. The length of the hydrogen bond is given in angstroms. B. Treatment with CPUY192018 disrupts KEAP1-PGAM5 interaction. PC6 cells were co-transfected with KEAP1-FLAG and PGAM5-HA and treated for 24 h with DMSO or 100 μM CPUY192018. KEAP1-FLAG was co-immunoprecipitating with PGAM5-HA in DMSO but not in CPUY192018 treated cells. C. Treatment with CPUY192018 increases the level of endogenous PGAM5. PC6 cells were treated with 100 μM CPUY192018 for 24 h. The left panel shows a representative Western blot and the right panel quantitative analysis. *P < 0.05, n = 4 samples, t-test. D. Effect of non-electrophilic KEAP1 inhibitors on mitochondrial membrane potential. PC6 cells were treated with KEAP1 inhibitors for 24 h, after which the cells were stained with ratiometric mitochondrial membrane potential sensor JC10. *P < 0.05 and ****P < 0.0001 when compared with DMSO treated group, n = 7–12 wells per data point, One-way ANOVA followed by Dunn's multiple comparisons test. E. Non-electrophilic KEAP1 inhibitors induce Parkin-EYFP translocation to mitochondria. PC6 cells expressing Parkin-EYFP were treated with DMSO or 100 μM of KEAP1 inhibitors for 24 h ***P < 0.001 and ****P < 0.0001 when compared with DMSO treated group, n = 6 dishes per group, 20 fields per dish, One-way ANOVA followed by Dunnett's multiple comparisons test. F and G. Non-electrophilic KEAP1 inhibitors enhance mitophagy but not general autophagy. Neurons expressing mitochondrial marker Kate2 and autophagosome markers EGFP-LC3B and GFP-LC3C were treated with DMSO or 100 μM of KEAP1 inhibitors for 24 h. The number of LC3 positive puncta colocalizing with mitochondria (F) as well as the total number of LC3 positive puncta (G) was counted by a blinded observer. **P < 0.01, ***P < 0.001 and ****P < 0.0001 when compared with DMSO treated group, n = 9–14 dishes (10 cells per dish), Welch's ANOVA followed by Dunnett's T3 multiple comparisons test. H. Treatment of primary cortical neurons with CPUY192018 (24 h) induces a concentration-dependent increase in mitochondrial colocalization with autophagosome markers EGFP-LC3B/GFP-LC3C. *P < 0.05 and ****P < 0.0001, when compared with DMSO, treated group, n = 16–18 dishes from 4 independent experiments (10 cells per dish), Kruskal-Wallis test followed by Dunn's multiple comparisons test, the dotted line shows the EC50.