Figure 2. Allele assignment by ‘default’, a commonly used strategy.
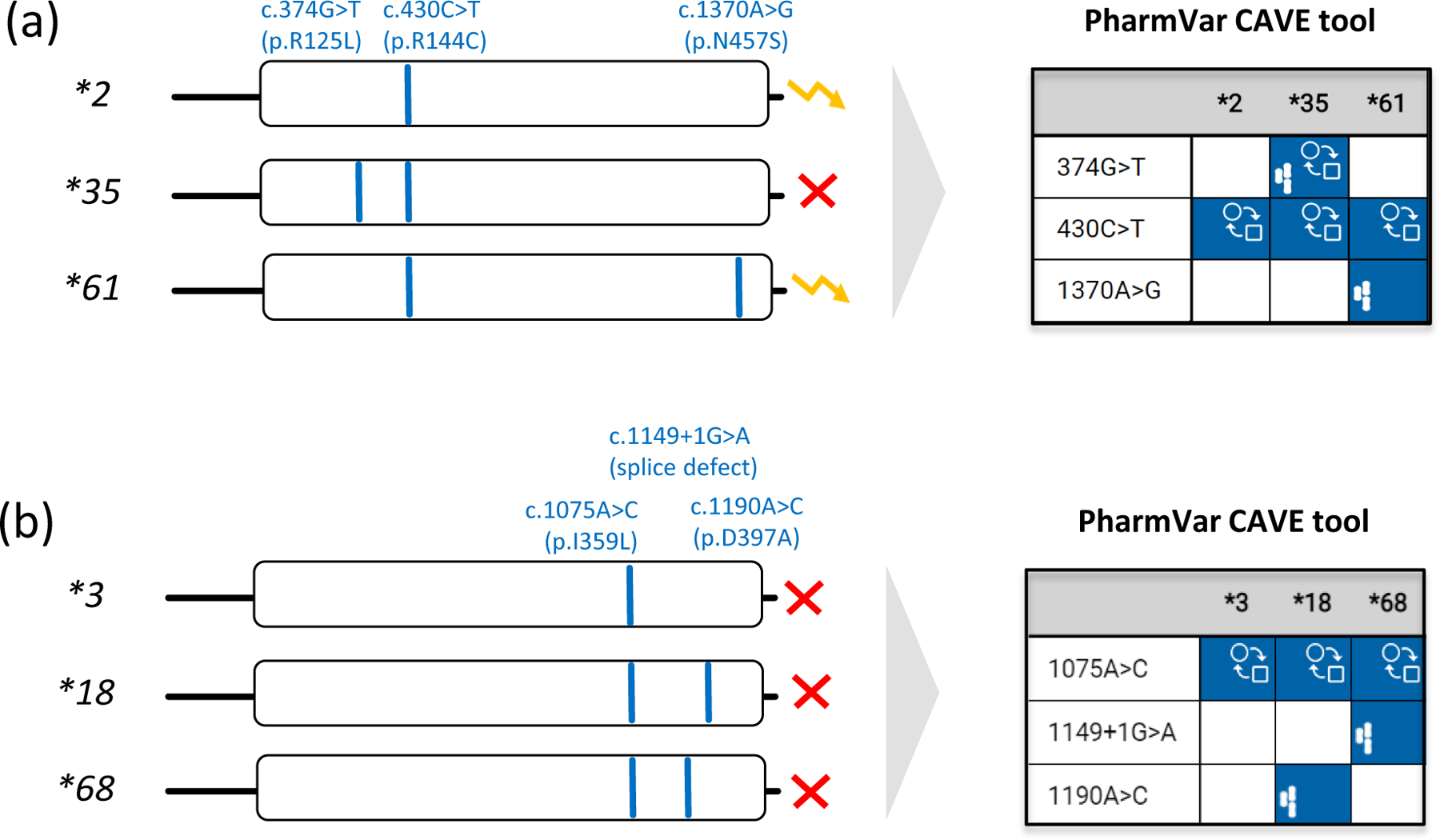
Pharmacogenetic test platforms typically comprise the more commonly observed SNVs and do not test for all star (*) alleles catalogued by PharmVar. Consequently, some alleles may not be identified or receive an assignment by ‘default’. Therefore, it is important to understand which SNVs are tested and how alleles were called and translated into phenotype. Panel (a) shows that the relatively common SNV at c.430C>T (p.R144C) is not only a core SNV of CYP2C9*2, but also *35 and *61. Unless c.374G>T and c.1370A>G are also tested, a CYP2C9*35 or *61 allele will be called as a *2 potentially leading to an incorrect phenotype assignment. Since CYP2C9*2 and *61 are both classified as decreased function by CPIC, defaulting CYP2C9*61 as *2 will not impact the patient’s phenotype call. Panel (b) demonstrates that c.1075A>C (p.I359L) is also present in CYP2C9*18 and the recently discovered *68 alleles. c.1075A>C on its own causes severely decreased function and thus, CYP2C9*3 is classified by CPIC as ‘no function’ allele. CYP2C9*68 has an additional SNV interfering with splicing predicted to be deleterious/probably damaging (93). It remains unknown though whether p.D379A on CYP2C9*18 also impacts function. The yellow ‘zigzag’ arrow and the red X denote decreased and no function, respectively.
Comparative
Allele
ViewEr (CAVE) outputs are shown for both examples to demonstrate the utility of this tool. The graph visualizes which core alleles have the tested SNVs and may be not be identified by limited testing and default allele assignments. Blue boxes indicate the presence of core SNVs. The function ( ) symbol indicates that a core SNV alters function and the PharmVar (
) symbol indicates that a core SNV alters function and the PharmVar ( ) symbol highlights that a core SNV is unique to a star allele. SNV positions refer to the transcript (NM_000771.4).
) symbol highlights that a core SNV is unique to a star allele. SNV positions refer to the transcript (NM_000771.4).
