Figure 4. Characterization of novel allelic variants.
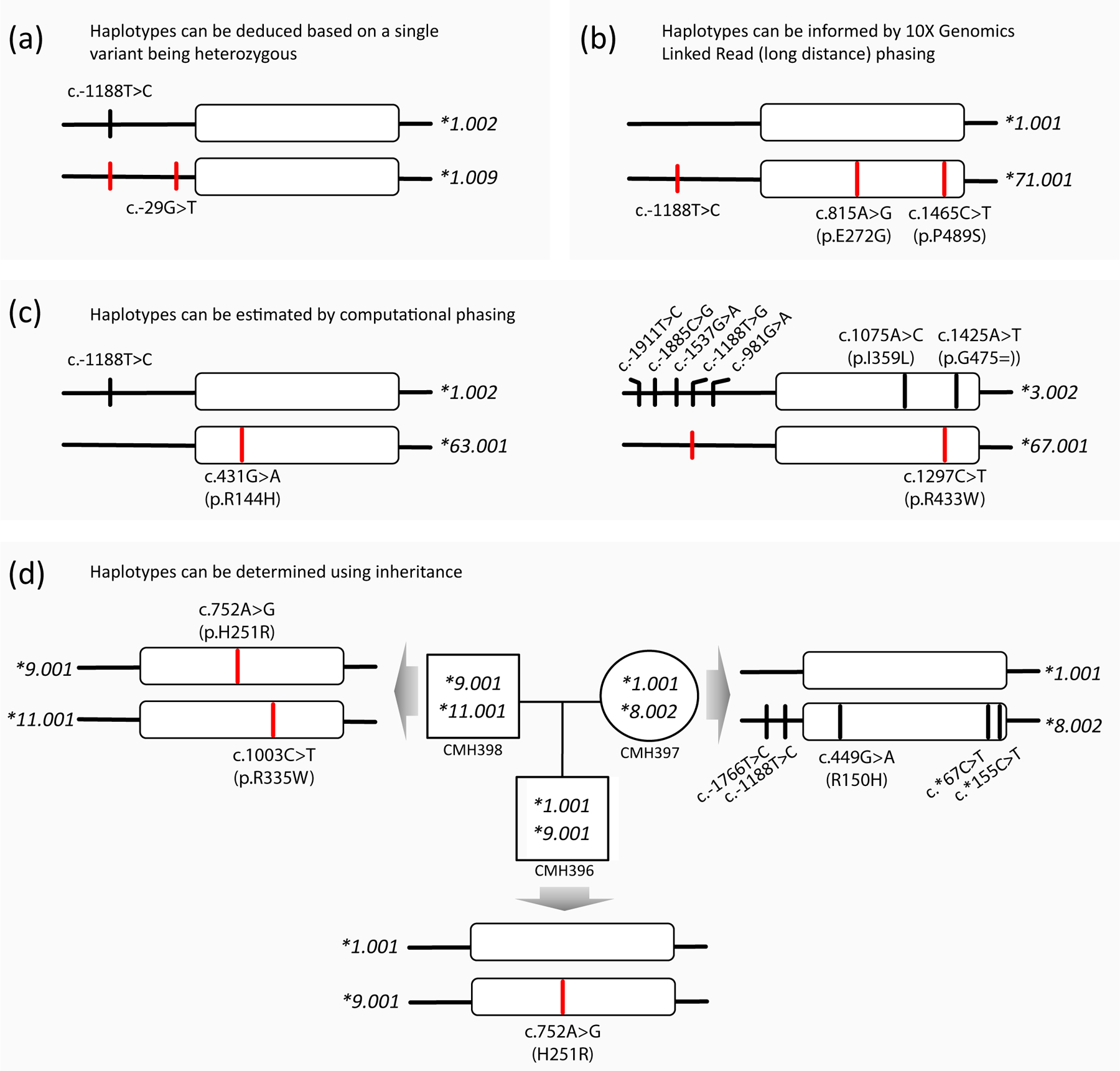
Panels (a-d) provide examples of alleles submitted to PharmVar for naming or to confirm existing allele definitions. Variants of submitted alleles are highlighted by red lines. All submissions utilized WGS data which were either confirmed by WES or targeted NGS-based sequencing panels. Panel (a) exemplifies a subject whose haplotype can be unequivocally be deduced and Panel (b) shows a subject whose three heterozygous SNVs were placed on the same chromosome using 10X Genomics Linked-Read (long-distance) phasing technology; CYP2C9*1.009 and *71 both received an evidence level of ‘Definitive’ (‘Def’). Panel (c) depicts two examples for which haplotypes were computationally inferred. Due to uncertainty regarding the phase of the variants, CYP2C9*63 and CYP2C9*67 received evidence levels of ‘Moderate’ (Mod) and ‘Limited’ (Lim), respectively. Panel (d) illustrates how haplotype can be inferred using inheritance in a family trio.
