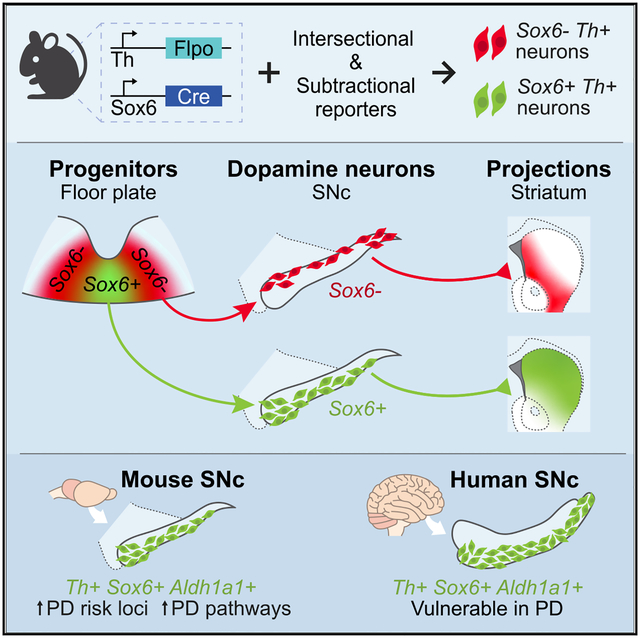
SUMMARY
Dopamine (DA) neurons in the ventral tier of the substantia nigra pars compacta (SNc) degenerate prominently in Parkinson’s disease, while those in the dorsal tier are relatively spared. Defining the molecular, functional, and developmental characteristics of each SNc tier is crucial to understand their distinct susceptibility. We demonstrate that Sox6 expression distinguishes ventrally and dorsally biased DA neuron populations in the SNc. The Sox6+ population in the ventral SNc includes an Aldh1a1+ subset and is enriched in gene pathways that underpin vulnerability. Sox6+ neurons project to the dorsal striatum and show activity correlated with acceleration. Sox6− neurons project to the medial, ventral, and caudal striatum and respond to rewards. Moreover, we show that this adult division is encoded early in development. Overall, our work demonstrates a dual origin of the SNc that results in DA neuron cohorts with distinct molecular profiles, projections, and functions.
Graphical Abstract
In brief
In this study, Pereira Luppi et al. refine the landscape of midbrain dopamine neurons through Sox6 lineage analysis to shed light on dorsal and ventral substantia nigra pars compacta from molecular, anatomical, functional, and developmental perspectives.
INTRODUCTION
The substantia nigra pars compacta (SNc), a key dopamine (DA) neuron containing cluster, is heterogeneous in its composition. Even before the discovery of (DA) 6 decades ago, Hassler demonstrated that in post-mortem Parkinson’s disease (PD) brains neurodegeneration is more prominent in the ventral SNc than in the dorsal SNc, a finding that has been corroborated using immunolabeling techniques for DA neuron markers (Damier et al., 1999; Giguère et al., 2018; Kordower et al., 2013; Liu et al., 2014; Mendez et al., 2005; Yamada et al., 1990) and has led to the description of a ventral and dorsal tier of the SNc. Uncovering the molecular, anatomic, and functional differences between these tiers would help explain their susceptibility in PD, yet they are still to be fully elucidated.
Besides distinctions based on selective vulnerability, recent studies suggest physiological and functional heterogeneity of SNc DA neurons. Physiologically, SNc neurons differ by their T type calcium channel-mediated rebound excitability (Evans et al., 2017), hyperpolarization-activated cyclic nucleotide-gated (HCN) channel current size (Lerner et al., 2015), or burstiness (Farassat et al., 2019). Functionally, some SNc neurons respond to rewarding stimuli, while others located in the lateral SNc or SN pars lateralis (SNpl) also respond to aversive stimuli (Lerner et al., 2015; Matsumoto and Hikosaka, 2009; Menegas et al., 2018). Axons of SNc neurons projecting to the dorsal striatum (DS) display activity time locked to locomotion, whereas those projecting ventrally appear to be more reward responsive (Howe and Dombeck, 2016). During movement, some are activated during acceleration, and some are unresponsive (Dodson et al., 2016; Howe and Dombeck, 2016; da Silva et al., 2018). These studies find major differences across the mediolateral (M-L) axis (Farassat et al., 2019; Lerner et al., 2015) or across the dorsoventral (D-V) axis (Evans et al., 2017) of the SNc. To better integrate and extend these findings, a deeper understanding of SNc heterogeneity is key to deciphering the pleiotropic functions of this structure.
To examine SNc heterogeneity at the molecular level, single-cell profiling has been used to group DA neurons based on similar molecular signatures. Recent studies have revealed the presence of several putative DA neuron subtypes (Hook et al., 2018; Kramer et al., 2018; La Manno et al., 2016; Poulin et al., 2014; Saunders et al., 2018; Tiklová et al., 2019). For instance, our lab and others have identified a DA neuron population defined by the expression of transcription factor Sox6 and aldehyde dehydrogenase Aldh1a1 (Panman et al., 2014; Poulin et al., 2014), located in the ventral SNc, that was selectively vulnerable in a toxin model of PD. Despite this promising start toward SNc classification, defining the complete catalog of the cellular constituents of the SNc remains a work in progress, in part because of the technical limitations of single-cell transcriptomic analyses, as well as the closely related nature of these neurons (Poulin et al., 2016). How these molecularly defined subtypes align with dorsal and ventral tiers remains to be fully addressed. Moreover, if there is such a fundamental difference in vulnerability, then it opens the possibility that it is encoded early in development.
All midbrain DA neurons originate from the Shh+/Foxa2+/Lmx1a+ embryonic floor plate (FP) (Andersson et al., 2006; Blaess et al., 2011; Joksimovic et al., 2009; Metzakopian et al., 2012; Yan et al., 2011), which can be subdivided roughly into a medial Sox6+ and a lateral Sox6−/Wnt1+/Otx2+ progenitor domain (Nouri et al., 2020; Panman et al., 2014; Prakash et al., 2006). Current models disagree on the molecular identity of the medial versus lateral progenitor domain descendants, with some studies suggesting that medially located progenitors represent a key anlage for SNc neurons (Blaess et al., 2011; Panman et al., 2014) and others suggesting that the lateral progenitor domain is also a prominent source of SNc neurons (Brown et al., 2011; Joksimovic et al., 2009). In addition, none of these studies have explained the developmental underpinnings of dorsal and ventral tier neurons. Using lineage tracing tools to understand SNc diversity from a developmental perspective could reinforce the robustness of adult single cell-based taxonomic dendrograms (Arendt et al., 2019; Fiorenzano et al., 2021).
This study aims to shed light on dorsal and ventral SNc DA neurons from molecular, functional, and developmental perspectives. First, we demonstrate that the SOX6+,ALDH1A1+ population exists in the human ventral SNc and is selectively diminished in post-mortem PD brains. Second, we reveal that Sox6 expression defines a population of neurons that are more ventrally located and enriched in molecular pathways that could underpin vulnerability. Furthermore, we uncover a dorsal Sox6− population that includes neurons expressing Calb1 and Slc17a6 (Vglut2), genes associated with resilience to neurodegeneration. We describe distinct projection patterns and functions of Sox6+ and Sox6− neurons, validating this fundamental SNc division. Moreover, we reveal distinct developmental histories between these neuronal cohorts that demonstrate a dual embryonic origin of the SNc. Our work facilitates a more granular definition of the cellular landscape of the SNc, beyond current schemes obtained by single-cell profiling studies, and provides insights into the selective vulnerability of ventral tier neurons in PD.
RESULTS
SOX6+,ALDH1A1+ DA neurons in the human ventral SNc are vulnerable in PD
Previous single-cell profiling analyses of murine SNc have identified a subset of DA neurons that co-expresses Sox6 and Aldh1a1 (La Manno et al., 2016; Poulin et al., 2014). This subset was selectively vulnerable in a mouse 1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine (MPTP) model (Poulin et al., 2014). To evaluate whether a similar neuron type exists in the human SNc (hSNc) and whether it is vulnerable in PD, we performed immunolabeling in control and PD post-mortem brains. In control brains, we observed a defined ALDH1A1+ subset localized ventrally in the hSNc (Figures 1A and 1B), the majority of which co-expressed SOX6+ (83.8% ± 3.4%; Figures 1C and S1B). To assess their vulnerability, we investigated PD brains and age-matched controls (Table S1). As expected, TH+ neurons were significantly reduced in PD brains (70.0% ± 4% reduction compared to control brains; Figures 1B and S1A), but most important, SOX6+,ALDH1A1+ DA neurons were more diminished in numbers than other DA neurons. TH+,SOX6+,ALDH1A1+ showed an 82.1% ± 3.7% reduction compared to controls, while other DA subtypes showed a 50.3% ± 6.8% reduction compared to controls (Figure 1D). To characterize resilient DA neuron cohorts, we stained sections for TH, CALBINDIN-D28k (CB), and ALDH1A1. Most CB+ DA neurons were ALDH1A1−, although a small subset expressed ALDH1A1 (Figures 1E and S1B). In PD brains, CB+ DA neurons were less affected than other DA neuron subtypes (TH+,CB+ 13% ± 13.9% reduction compared to controls, other DA subtypes 69.4% ± 4.4% reduction compared to controls; Figure 1F). In conclusion, our experiments showed that a selectively vulnerable population in the ventral hSNc co-expresses SOX6 and ALDH1A1.
Figure 1. SOX6+,ALDH1A1+ neurons in human SNc are vulnerable in PD while CALBINDIN-D28k+ neurons are relatively spared.
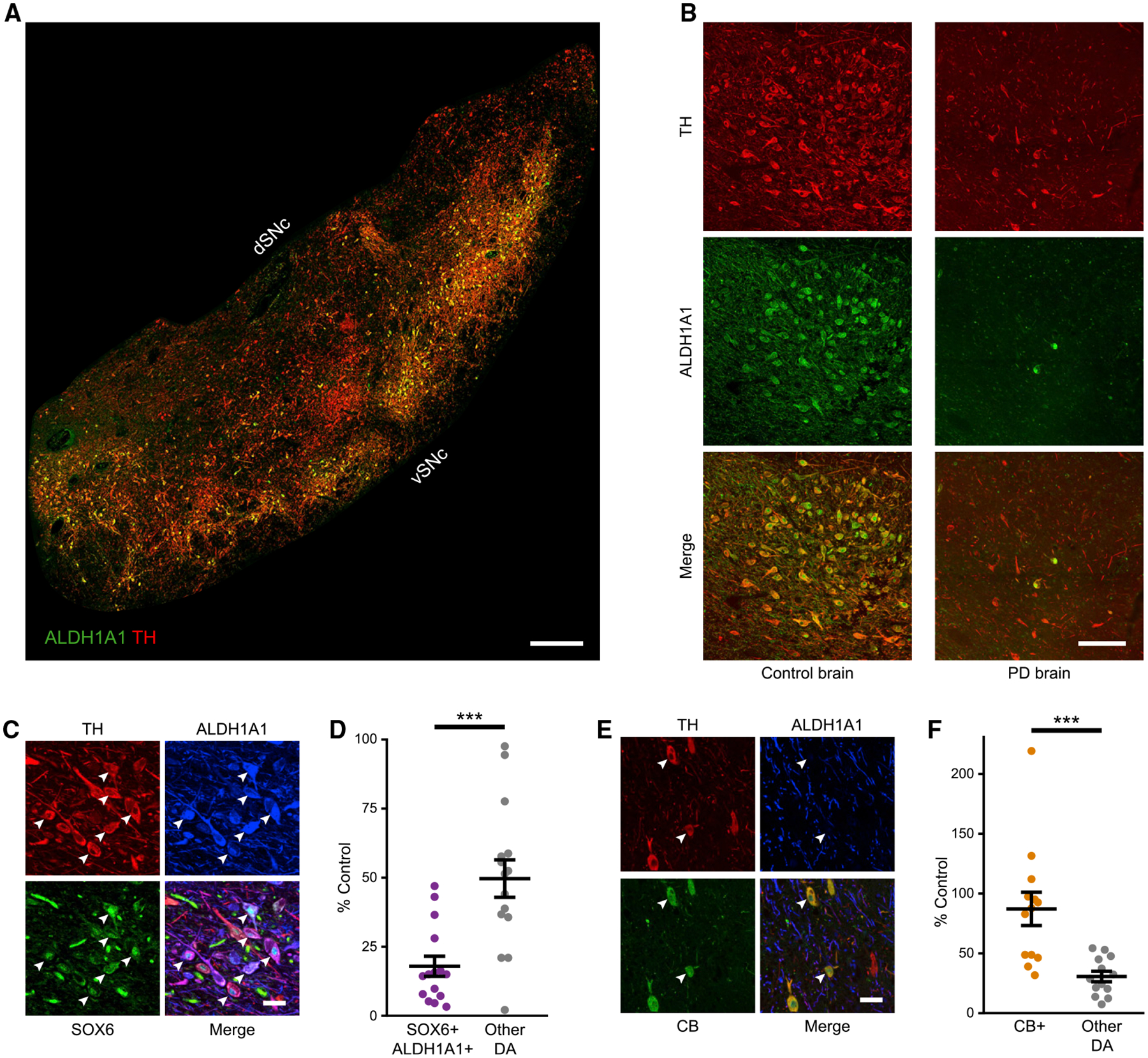
(A and B) TH+,ALDH1A1+ labeling in control and PD brains. In PD brains, ALDH1A1 staining appears brighter in the nuclei compared to controls, as previously described in a 6-OHDA model (Stott and Barker, 2014).
(C) High magnification of SOX6+,ALDH1A1+ DA neurons (arrowheads) in a control brain.
(D) SOX6+,ALDH1A1+ DA neurons versus other DA neurons in hSNc of PD as a percentage of control brains (p = 3.1 × 10−4; controls are set at 100%, n = 14 controls, n = 15 PD).
(E) High magnification of CALBINDIN-D28k+ (CB+) DA neurons in a control brain.
(F) CB+ DA neurons versus other subtypes in PD as a percentage of control brains (p = 7.3 × 10−4, n = 14 controls, n = 15 PD).
Scale bars: (A) 1,000 μm, (B) 100 μm, and (C and E) 50 μm. Error bars are SEMs. vSNc, dSNc: ventral and dorsal substantia nigra pars compacta. hSNc: human substantia nigra pars compacta.
Sox6 expression distinguishes dorsally and ventrally biased cohorts in the murine SNc
Sox6 expression is a primary factor that distinguishes DA neurons (La Manno et al., 2016; Panman et al., 2014; Poulin et al., 2014), making it an excellent genetic entry point to understand DA neuron diversity in mice. Sox6 is widely expressed in SNc DA neurons (Panman et al., 2014; Poulin et al., 2014), but also in DA neuron progenitors (Panman et al., 2014), GABAergic neurons, and glia intermingled and in the vicinity of the SNc. Therefore, to specifically visualize post-mitotic Sox6+ and Sox6− DA neurons, we developed a simultaneous intersectional/subtractive strategy using our validated lines Sox6-FSF-Cre and Th-2A-Flpo. In this model, Cre activity is restricted to Sox6+,Th+ neurons. When combined with a RC-Frepe reporter, Sox6−,Th+ neurons (Cre−, Flpo+) express mCherry while Sox6+,Th+ neurons (Cre+, Flpo+) express GFP (Figure 2A). In the SNc of Sox6-FSF-Cre, Th-2A-Flpo, RC-Frepe adult brains, most DA neurons were GFP+ (70.0% ± 3.1%; Figures 2B and 2C). However, we observed a clear population of mCherry+ DA neurons, indicating either insufficient levels or complete lack of Sox6 expression. We also observed a small subset of mCherry+ neurons within the boundaries of the SNc, especially at the dorsal edge, that lacked TH immunolabeling (12.9% ± 0.01% of the SNc total mCherry+ population), in addition to mCherry+,TH− neurons above the SNc (Figure 2B).
Figure 2. Sox6 expression defines a dorsal-ventral division of the murine SNc.
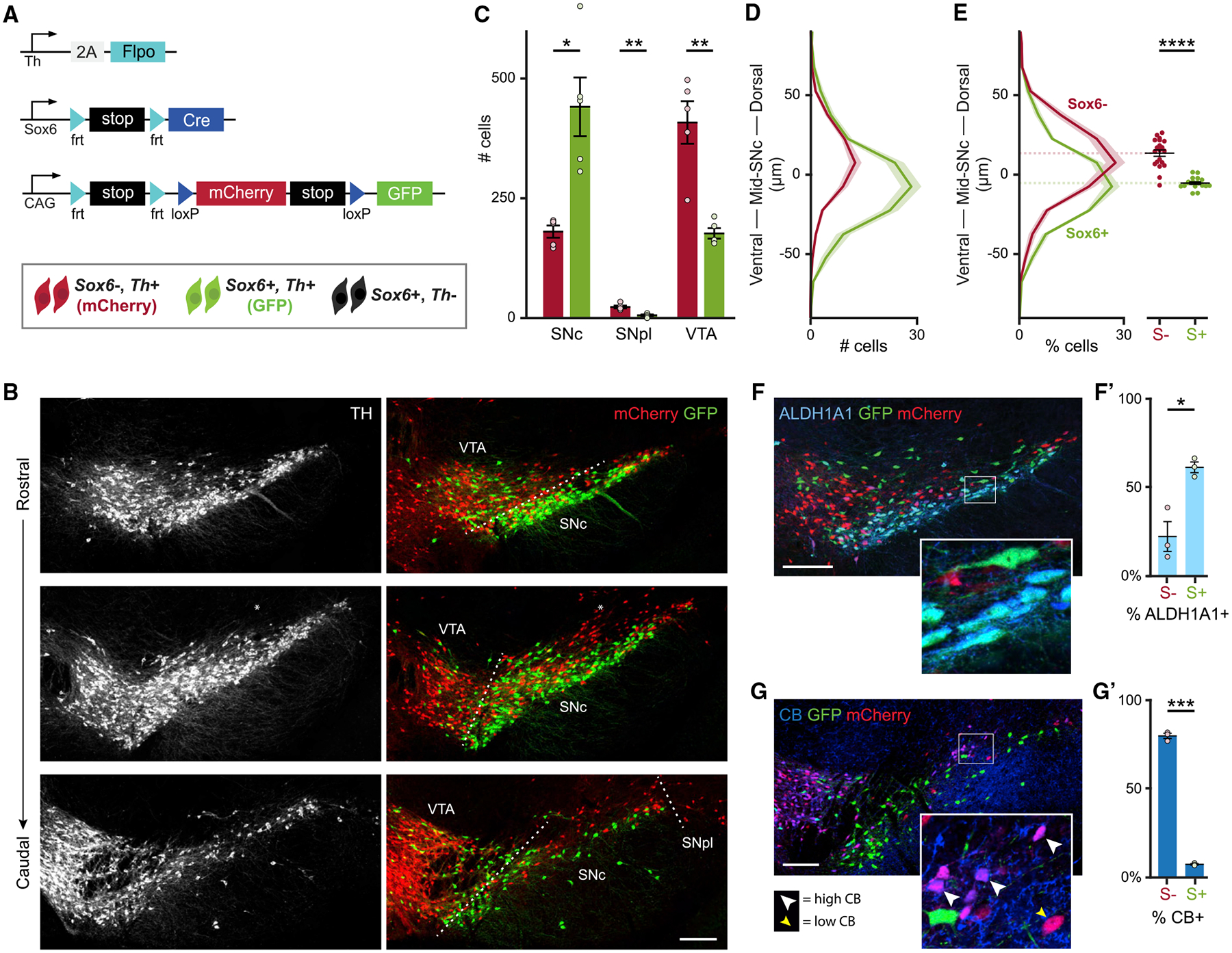
(A) Simultaneous intersectional/subtractional genetic labeling strategy.
(B) Example of an adult Sox6-FSF-Cre, Th-2A-flpo, RC-Frepe brain. Some mCherry+ cells are observed above the SNc, lacking TH immunoreactivity (*).
(C) Average number of TH+,mCherry+ (red bars) and TH+,GFP+ (green bars) in the main DA neuron areas. p values: SNc = 0.014, SNpl = 0.004, VTA = 0.008.
(D) Histogram of spatial coordinates of TH+,GFP+ or TH+,mCherry+ cells; TH+,mCherry+ neurons are biased dorsally.
(E) Left: histogram of spatial coordinates of TH+,GFP+ or TH+,mCherry+ cells normalized to total TH+,GFP+ or TH+,mCherry+ cells. Right: centroid of GFP+ and mCherry+ populations for each section of each mouse included in left panel (4 sections per brain) (p = 6 × 10−7).
(F) GFP+ cells co-express ALDH1A1 (SNc).
(F’) Percentage of GFP+ (Sox6+) and mCherry+ (Sox6−) neurons co-expressing ALDH1A1 in the SNc. p = 0.02.
(G) Most mCherry+ cells express CALBINDIN-D28k (CB) (SNc). CB signal intensity varies from low (yellow arrowhead) to high (white arrowhead).
(G’) Percentage of GFP+ (Sox6+) and mCherry+ (Sox6−) neurons co-expressing CB in the SNc. p = 0.0007.
Scale bars: 200 μm. Error bars (C’, F’, and G’) and shaded areas (D and E) are SEMs. S+: Sox6+, S−: Sox6−. (A)–(E) n = 5, (F) and (G) n = 3.
Remarkably, reporter-labeled cells displayed a biased spatial distribution: GFP+ neurons appeared to be ventrally enriched and mCherry+ neurons appeared to be dorsally enriched, although there was significant intermingling (Figures 2B and 2D). We took two approaches to quantify the spatial distribution of TH+,GFP+ and TH+,mCherry+ cells. First, we bisected the SNc into a ventral half and a dorsal half (see Method details). We found GFP+ DA neurons approximately evenly distributed between these halves, whereas 70.4% ± 4.3% of mCherry+ DA neurons were located in the dorsal half (data not shown). Second, we plotted the spatial coordinates of SNc TH+,mCherry+ and TH+,GFP+ DA neurons relative to line of best fit given all SNc cells (Figures S2A–S2A”‘). A histogram of the coordinate distance to the reference line showed that mCherry+ neurons were biased dorsally, while GFP+ neurons were distributed mainly ventrally but also intermingled with mCherry+ neurons in the dorsal SNc (Figure 2D). Since the mCherry+ population had fewer cells than the GFP+ population, we normalized the counts and observed a statistically significant difference in the centroids of the spatial distribution of GFP+ and mCherry+ neurons (Figure 2E). Finally, along the rostrocaudal axis of the SNc, our analysis showed that GFP+ neurons were more abundant than mCherry+ neurons, and that the latter were biased dorsally at all section levels (Figures S2B and S2C).
Other DA neuron clusters also exhibited a distinct proportion of reporter-labeled DA neurons: the SNpl appeared contiguous with the dorsal mCherry+ cells and was also predominantly mCherry+ (81.8% ± 6.0%; Figures 2B and 2C). In the ventral tegmental area (VTA), most DA neurons expressed mCherry (69.0% ± 3.0%; Figures 2B and 2C), indicating that they lacked or had insufficient levels of Sox6 expression. Nonetheless, we observed a GFP+ population in the VTA, located mainly in the parabrachial region. The retrorubral field (RRF), however, showed an even proportion of GFP+ and mCherry+ neurons, with significant intermingling (Figures S2D and S2E). In the interfascicular, paranigral, rostral linear, and caudal linear nuclei, the GFP+ population was less abundant (not shown).
To investigate whether the Aldh1a1+ population is fully or partially included in the Sox6+ (GFP+) population, we examined Aldh1a1 expression in Sox6-FSF-Cre, Th-2A-Flpo, RC-Frepe adult brains. In the SNc, ALDH1A1+ neurons, ventrally located, were mostly embedded in the GFP+ population (Figure 2F). Specifically, 86.7% ± 6.4% of the ALDH1A1+ neurons were GFP+, consistent with our hSNc data (Figure S1B). Furthermore, 61% of the GFP+ cells in the SNc were ALDH1A1+ (Figure 2F’), indicating the presence of both Sox6+,Aldh1a1+ and Sox6+,Aldh1a1− populations, in line with previous studies (Poulin et al., 2014).
Since Sox6 and Calb1 are key drivers of variance in single-cell studies (Poulin et al., 2014), we performed CB staining in Sox6-FSF-Cre, Th-2A-Flpo, RC-Frepe brains. CB+ neurons localized dorsally in the SNc, with high and low immunolabeling levels, as previously shown (Gerfen, 1985; La Manno et al., 2016) (Figure 2G). Remarkably, we found that the majority of mCherry+ neurons in the SNc co-expressed CB (79.3%; Figure 2G’). This was also the case in the VTA, 87.2% ± 2.1%, and the SNpl, 90.3% ± 6.6% (data not shown). Our findings strongly suggest that Sox6 expression divides the murine SNc into a Sox6+ population biased to the ventral SNc (vSNc) and a Sox6− population biased to the dorsal SNc (dSNc) and SNpl. In addition, Aldh1a1+ neurons are largely included in the Sox6+ vSNc, and a substantial portion of the Sox6− dSNc/SNpl expresses Calb1.
Sox6+ and Sox6− neurons display large transcriptomic differences, some of which may underpin selective vulnerability
To determine the molecular differences between ventral Sox6+ and dorsal Sox6− DA neurons, we performed bulk RNA sequencing (RNA-seq) in sorted mCherry+ and GFP+ cells from the SNc of adult Sox6-FSF-Cre, Th-2A-Flpo, RC-Frepe mice. Our analysis revealed large and significant differential gene expression between these two populations (Figures 3A and 3B; Table S2). Consistent with our recombinase-based labeling strategy and our previous results (Figure 2), Sox6 and Aldh1a1 were preferentially expressed in GFP+ neurons. In addition, genes likely to confer distinctive properties on neurons, such as Drd2, Vmat2, Kcnj6, and Kcns3, were significantly enriched in the GFP population, whereas others such as the ion transport regulator Fxyd6, neuroglobin (Ngb), and Slc17a6 (Vglut2) were enriched in the mCherry population (Figure 3A). In situ hybridization of Fxyd6, Ngb, and Vglut2 confirmed their expression in mCherry+,TH+ neurons in the dSNc and SNpl (Figure S3A). These markers were also expressed in mCherry+,TH− neurons located above the TH+ SNc. In contrast, somatostatin (Sst) was differentially expressed in the mCherry population, but was found only in mCherry+,TH− neurons above the SNc, indicating that some of these cells were included in our transcriptomic analysis (Figure S3A). These mCherry+,TH− neurons, like the TH+ cohorts, have a history of Th expression, and they are FOXA2+ indicative of a FP origin (data not shown). We postulate that these cells are related to dSNc DA neurons but fail to initiate or maintain robust expression of the DA program, and are therefore TH−.
Figure 3. Transcriptomic analysis of Sox6+ and Sox6− SNc cohorts.
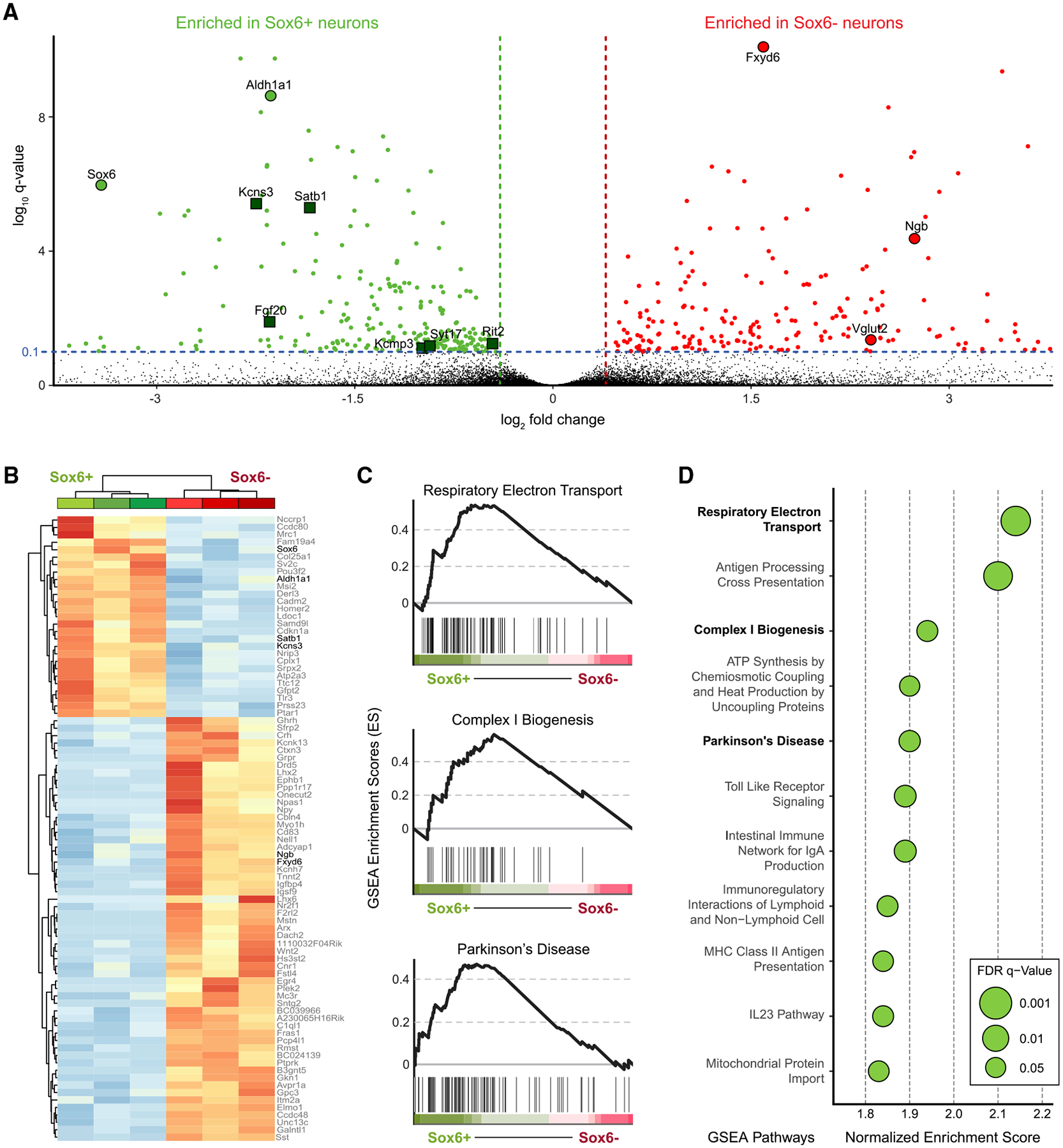
(A) Volcano plot showing gene expression differences between GFP+ (Sox6+) and mCherry+ (Sox6−) cells. Green squares are PD risk loci.
(B) Unbiased clustering of Sox6+ and Sox6− samples, n = 3 each.
(C) Gene set enrichment analysis (GSEA) plots of gene sets enriched in the Sox6+ population associated with mitochondrial function and ATP synthesis (top, center) and PD (bottom). The peak of the curve indicates the enrichment score (ES), black lines below the curve, to the left of the peak, indicate the genes contributing the most to the peak (leading edge subset). q < 0.05 in all plots.
(D) Dot plot of GSEA ES in pathways enriched in Sox6+ neurons.
Our results indicated that the Sox6+ population includes Aldh1a1+ DA neurons (Figures 2F and 3A), which are vulnerable in PD models; therefore, we asked whether PD risk loci identified by genome-wide association studies (GWASs) (Nalls et al., 2019) were differentially expressed in the Sox6+ population. Interestingly, we found that six PD loci were enriched in the GFP, but none were enriched in the mCherry cohort (false discovery rate [FDR] < 0.1; green squares, Figure 3A). Consistent with this, when we performed unbiased pathway enrichment analysis using gene set enrichment analysis (GSEA), a PD-linked gene set was differentially expressed in GFP+ neurons (4th highest enrichment score [ES], q = 0.04; Figures 3C and 3D). In total, our GSEA analysis revealed 11 pathways differentially enriched in the GFP population (q < 0.05), some of which appear to be highly relevant to existing theories of PD pathogenesis (Surmeier, 2018). For example, three of these pathways were relevant to mitochondrial electron transport and ATP synthesis (electron transport and complex I biogenesis; Figures 3C, 3D, and S3B). Several immune-related pathways were also differentially enriched in GFP+ neurons, including innate immunity and Toll-like receptor (TLR) signaling (Figure S3B), which have been of recent interest in PD research (Cebrián et al., 2014; Dzamko et al., 2017).
Our simultaneous intersectional/subtractive strategy to access Sox6+ and Sox6− SNc populations allowed us to better interpret available single-cell sequencing datasets. We plotted differentially expressed genes using the DropViz platform (Saunders et al., 2018) (clusters 4–1 to 4–9 represent DA neurons; Figure S4). Interestingly, Sox6 expression discriminates between clusters 4–3, 4–5, 4–6, 4–7, 4–8, and 4–9 (Sox6+) and 4–1, 4–2, and 4–4 (Sox6−), consistent with our previous findings showing Sox6 expression as a key driver of variance (Poulin et al., 2014). Accordingly, we hypothesized that clusters 4–3, 4–5, 4–6, 4–7, 4–8, and 4–9 represent the GFP population. Aldh1a1, Kcns3, Atp2a3, Serpine2, and Satb1, significantly enriched in Sox6+ neurons, were highly expressed in these clusters. Conversely, clusters 4–1, 4–2, and 4–4 expressed a combination of Calb1, Vglut2, and Fxyd6, which we have shown to be enriched in Sox6− neurons (Figures 2G, 3A, and S3A), suggesting these could represent the mCherry population. Thus, we hypothesize that clusters 4–1, 4–2, and 4–4 represent together the VTA, dSNc, and SNpl, all predominantly Sox6− by our previous analysis (Figure 2). We attempted to match Sox6− clusters with these midbrain areas and found that Otx2 and Lpl, known ventromedial VTA markers (Poulin et al., 2020a), were practically exclusive to cluster 4–1. Calb1 was more widely expressed, in accordance with our results (Figure 2G), although more prominently in the Sox6− clusters, consistent with our previous study (Poulin et al., 2014). Thus, cluster 4–2, expressing Calb1, Fxyd6, and Vglut2, but not Otx2 and Lpl, can be associated with dSNc and SNpl, predominantly mCherry+ regions. All in all, Sox6− (mCherry+) cells are similar in that they lack Sox6, but express Calb1, Fxyd6, and/or Vglut2. However, Otx2 and Lpl allow us to distinguish between dSNc/SNpl (4–2; 4–4) and VTA (4–1).
In conclusion, our results provide direct evidence of substantial molecular distinctions between Sox6+ and Sox6− neurons, and that Sox6+ neurons are enriched in known pathways of DA neuron vulnerability. Moreover, the transcriptomic dataset derived from our intersectional/subtractive labeling scheme provides an informative resource, readily combinable with single-cell transcriptomics analyses for a more granular classification of DA neurons such as localizing neurons of the dSNc/SNpl.
Sox6+ and Sox6− neurons have distinct projections and functional responses
Our intersectional/subtractive labeling approach (Figure 2) labels Sox6+ and Sox6− DA neurons in the SNc but also in VTA and RRF, thus confounding analysis of the axonal projections of SNc neurons. To establish the projection targets of nigral Sox6+ and Sox6− neurons, we used an intersectional/subtractional viral strategy (Figures 4A and 4B) that labeled mainly SNc cells, with few VTA cells labeled (Figures 4D and S5B). Adult SNc of Sox6-FSF-Cre, Th-2A-Flpo mice were injected with CreOn/FlpOn-EYFP and CreOff/FlpOn-mCherry viruses, simultaneously labeling Sox6+ and Sox6− cohorts. Projections from Sox6+ and Sox6− populations targeted distinct areas of the caudo-putamen (CP) along the M-L, D-V, and rostro-caudal (R-C) axes (Figures 4C, S5C, and S5F). mCherry fibers from Sox6− neurons densely innervated the CP mainly in the medial and ventral areas, including the nucleus accumbens (ACB), and the tail of the striatum (TS) (Figure 4C). In rostral (CPr) and intermediate (CPi) sections, mCherry fibers were denser toward ventral areas, and in DS, they were strongly biased toward the medial striatum (Figure 4F). Conversely, EYFP fibers (Sox6+) densely innervated the DS, but showed no obvious bias along the M-L axis, unlike mCherry axons (Figures 4C and 4F). Labeled fibers were sparse in the ACB, except in parts of the core and lateral shell (Figure 4C). In more caudal sections (CPc), the D-V and M-L gradients become less prominent (Figure 4C). mCherry innervation is also particularly prominent in the interstitial nucleus of the posterior limb of the anterior commissure (IPAC) (Figures 4C and S5), in agreement with a previous report (Yamaguchi et al., 2018). To rule out biases from virus and/or fluorophore distribution, we repeated these experiments with a different set of viruses that label the same populations in reverse colors, with similar results (Figures S5D–S5F).
Figure 4. Sox6+ and Sox6− populations have distinct and inverted projection patterns.
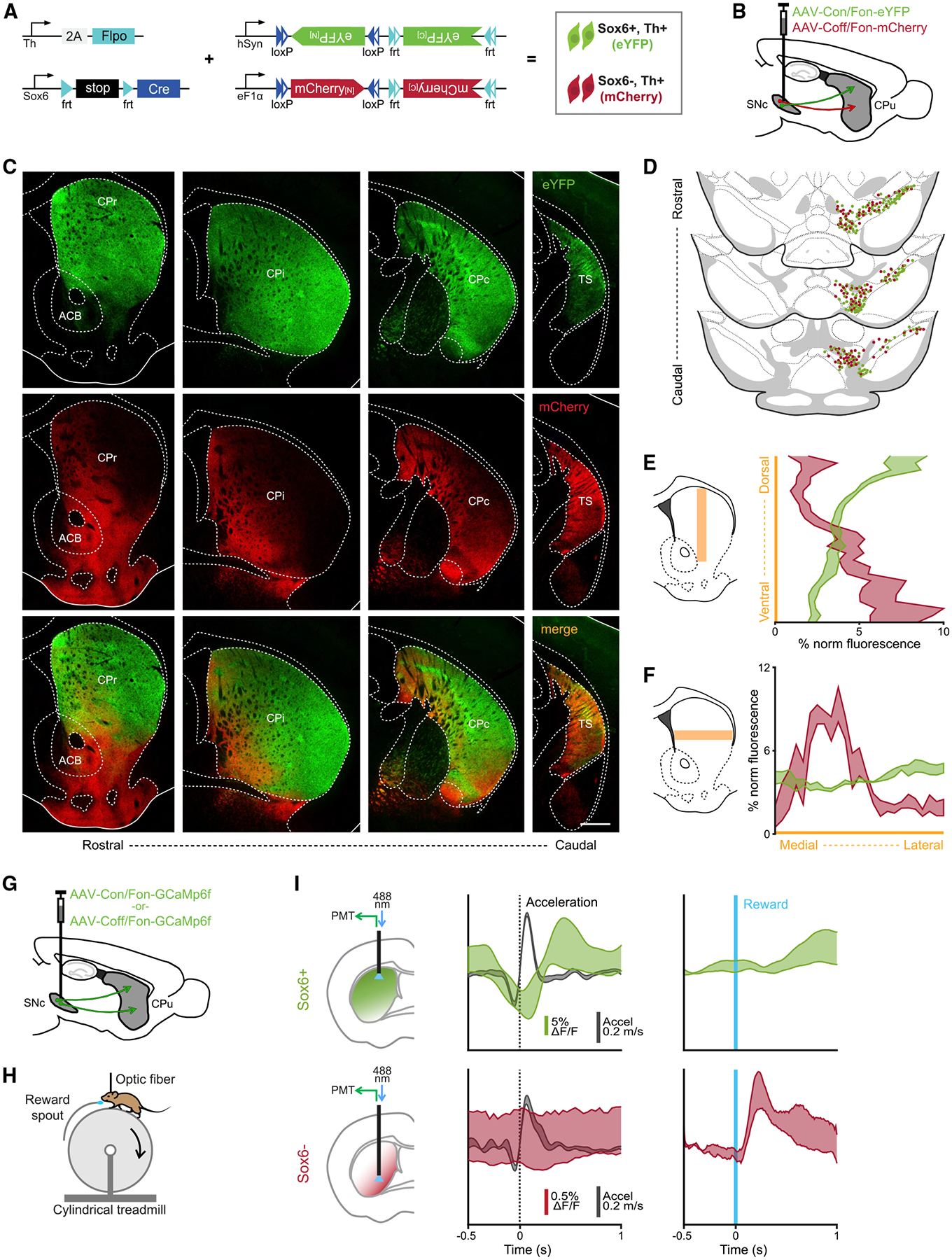
(A) Genetic strategy used to simultaneously label Sox6+ and Sox6− neurons and their projections in adult brains.
(B) Schematic showing injection location (SNc).
(C) Projections across the rostro-caudal axis of the striatum from Th+Sox6+ dSNc neurons (EYFP, top), from Th+Sox6− vSNc neurons (mCherry, center), and merged images (bottom).
(D) Representation of labeled cells in the midbrain corresponding to (C). Each dot represents 3 labeled cells.
(E) Histogram of normalized fluorescence for EYFP and mCherry axons along the dorsoventral axis of the striatum (CPr and CPi). Left, representation of the region of the striatum used for quantification.
(F) Histogram of normalized fluorescence for EYFP and mCherry axons along the mediolateral axis of the striatum (CPr and CPi). Left, representation of the region of the striatum used for quantification.
(G) Schematic showing strategy used to label Sox6+ or Sox6− neurons in different mice with GCaMP6f.
(H) Simplified representation of experimental setup.
(I) Recording of Sox6+ (top) and Sox6− (bottom) axons in the dorsal and ventral striatum, respectively in the regions of most dense innervation for each population. Left: schematic of recording location for each population. Center: triggered average at acceleration onsets (acceleration in black, %ΔF/F in red/green) (Sox6+ n = 9, Sox6− n = 2). Right: triggered average at reward delivery, %ΔF/F scale same as in the center (Sox6+ n = 6, Sox6− n = 2).
Shaded areas (E, F, I) are SEMs. Scale bar, 500 μm. CPr: caudate putamen rostral; Cpi: caudate putamen intermediate; CPc: caudate putamen caudal; TS: tail of the striatum; ACB: nucleus accumbens.
(A)–(F) n = 3.
Because of the anatomical proximity of the SNc and VTA, it is technically challenging to label all of the SNc while avoiding all VTA cells. To confirm that the projection patterns observed were not due to the VTA cells labeled, we repeated the experiment with a smaller, more lateral viral injection, which spared the VTA but failed to fully label the medial SNc (Figures S5A–S5C). While the projection patterns were similar as shown in Figure 4C, projections into the ACB core and olfactory tubercle were absent in these mice (Figures S5A–S5C). In the dorsal CP, EYFP axons were sparser in medial regions, possibly due to medial SNc cells not labeled in this strategy (Figures S5A–S5C), in accordance with Lerner et al. (2015). Overall, the Sox6+ projection pattern agrees with our previous work (Poulin et al., 2018). Here, we additionally show that the Sox6− population of the dSNc/SNpl has markedly distinctive projections, including more densely to the medial, ventral, and tail of the striatum. This roughly matches the projections of Calb1+ neurons, consistent with the fact that Sox6− neurons are largely Calb1+ (Figure 2G).
A previous study reported a subset of DA axons with fast signaling associated with locomotion in the dorsal CP (Howe and Dombeck, 2016). This motor-associated DA signaling was found in DS projecting axons, while more ventral axons responded to rewards. Combined with our anatomical observation, this suggests that these motor-associated dopamine neurons would be included in the Sox6+ population. To confirm this, we used fiber photometry to record calcium transients on genetically defined axons in behaving mice. We labeled Sox6+ or Sox6− SNc DA neurons with calcium indicator GCaMP6f by injecting Sox6-FSF-Cre, Th-2A-Flpo mice with either an intersectional (AAV-COn/FOn-GCAMP6f) or subtractional (AAV-COff/FOn-GCaMP6f) virus (Figure 4G). Following viral injections, head-restrained mice received unexpected random water rewards while running freely on a cylindrical treadmill (Howe and Dombeck, 2016) (Figure 4H), at the same time we recorded calcium transients using one-photon fiber photometry. Recordings from Sox6+ axons in DS showed calcium transients time locked to acceleration and no detectable response to rewards, as expected (Howe and Dombeck, 2016) (Figure 4I, top), confirming that the Sox6+ population includes motor-associated DA neurons. This does not, however, exclude the possibility of additional diversity within Sox6+ neurons (Poulin et al., 2020a). Recording from Sox6− axons in the CP proved more challenging (particularly in mid-depth), due to labeling inefficiencies of the available virus, as well as the lower density of Sox6− axons in the CP. However, in two mice, we observed reward-locked calcium transients in ventral CP axons of Sox6− neurons (Figure 4I, bottom right). As expected, in these same recordings, we did not detect any acceleration-locked calcium transients (Figure 4I, bottom).
Sox6+ and Sox6− neurons have distinct developmental histories
Sox6 expression is restricted to a subset of DA progenitors (Panman et al., 2014). Thus, we investigated whether differences observed between Sox6+ and Sox6− neurons could be encoded early in development. To this end, we designed a multi-pronged series of lineage tracing experiments to capture the full history of Sox6 expression from the progenitor stage.
We analyzed Sox6-FSF-Cre, CAG-Flpe, RC-Frepe embryos at embryonic day (E)11.5 and observed GFP+ cells located medially in the Lmx1a+ domain (Figure 5A), consistent with Sox6 embryonic expression (Panman et al., 2014). This result confirmed that our strategy appropriately labeled progenitors in the midbrain FP. Thus, we obtained a Sox6-Cre mouse line from our existing Sox6-FSF-Cre line, by permanently deleting the frt-flanked stop cassette with CAG-Flpe (Figure S6A). To create a cumulative fate map of Sox6 while simultaneously labeling Sox6+ and Sox6− populations, we crossed Sox6-Cre to Th-2A-Flpo and RC-Frepe. In Sox6-FSF-Cre animals (Figure 2), the intersectional reporter RC-Frepe labels only postmitotic cells expressing Sox6, whereas in Sox6-Cre animals, Cre recombination in RC-Frepe can occur in both progenitors and post-mitotic neurons, labeling with GFP all of the neurons that have expressed Sox6 at any point in their history and with mCherry those DA neurons that have never expressed sufficient levels of Sox6 (Figure 5D).
Figure 5. Sox6 cumulative fate map.
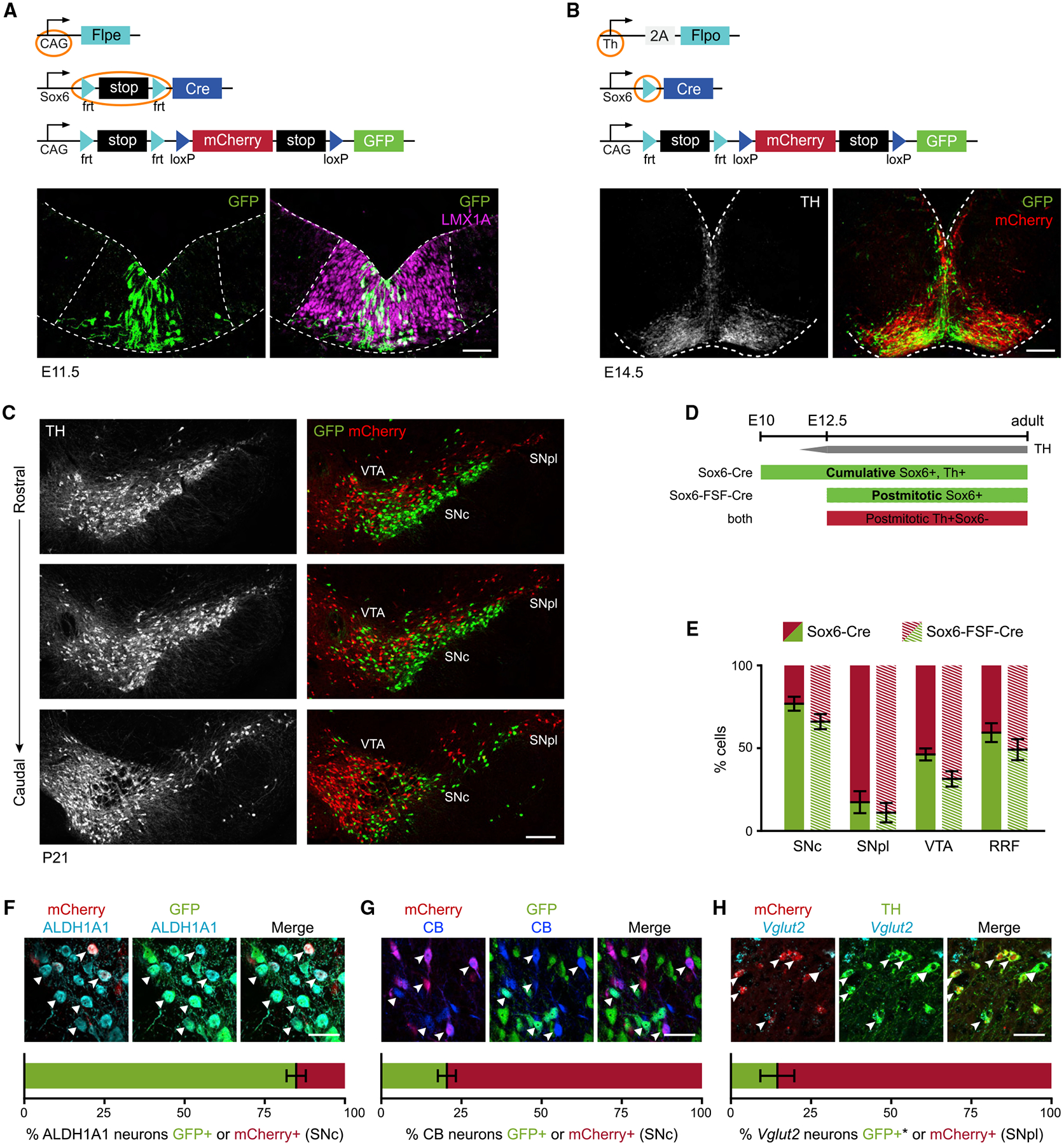
(A) Sox6-FSF-Cre, CAG-Flpe, RC-Frepe brain at E11.5 (mCherry not shown).
(B) Sox6-Cre, Th-2A-Flpo, RC-Frepe brain at E14.5.
(C) Adult Sox6-Cre, Th-2A-Flpo, RC-Frepe brain (n = 9).
(D) In Sox6-FSF-Cre, Th-2A-Flpo, RC-Frepe brains, DA neurons expressing Sox6 postmitotically (after the onset of Th expression) are labeled with GFP. In Sox6-Cre, Th-2A-Flpo, RC-Frepe brains, Cre activation no longer depends on Th; thus, TH+ DA neurons expressing Sox6 at any point in their history are labeled with GFP.
(E) Comparison of cumulative versus postmitotic labeling. In VTA, there are more GFP+ neurons in the cumulative analysis (p = 0.06).
(F and G) SNc of Sox6-Cre, Th-2A-Flpo, RC-Frepe brains labeled with ALDH1A1, or CALBINDIN-D28k+ (CB+) (GFP, triangles; mCherry arrowheads).
(H) In the SNpl, the majority of Vglut2+ neurons are TH+,mCherry+ (arrowheads), while only a few are TH+,mCherry− (triangles). *Typically GFP+, lost during in situ hybridization.
Scale bars: (A) 50 μm; (B) 100 μm; (C) 200 μm. Error bars are SEMs. (A, B, and E–G) n = 3.
We examined Sox6-Cre, Th-2A-Flpo, RC-Frepe embryos at the end of DA neurogenesis, E14.5 (Figure 5B). In these brains, GFP+ and mCherry+ cells had a distinct distribution as they emanated from the FP into the mantle zone. GFP+ cells were densely located medially, likely migrating radially and then tangentially at the pial surface as previously reported (Bodea et al., 2014; Panman et al., 2014; Vaswani et al., 2019). mCherry+ cells were prominent laterally, with some GFP+ cells intermingled. Interestingly, we observed a few mCherry+,GFP+ cells, also seen in E16.5 embryos, indicating that some DA neurons activate Sox6 expression postmitotically (Figure S6B).
To determine the contribution of Sox6 expression to the adult midbrain, we analyzed Sox6-Cre, Th-2A-Flpo, RC-Frepe brains. In the SNc, we observed a prominent ventral TH+,GFP+ population and a less abundant dorsal TH+,mCherry+ population (Sox6+ and Sox6− history, respectively; Figure 5C). Conversely, the SNpl was predominantly populated by TH+,mCherry+ neurons. We compared the percentage of GFP and mCherry-expressing cells in Sox6-Cre versus Sox6-FSF-Cre animals and found no statistically significant difference in SNc, SNpl, and RRF. However, there were more GFP+ cells in the VTA of Sox6-Cre animals compared to Sox6-FSF-Cre (p = 0.06; Figure 5E). In fact, in Sox6-FSF-Cre brains, we observed mCherry+ cells throughout the VTA with some GFP+ cells in the parabrachial region (Figure 2B), whereas in Sox6-Cre brains, we also observed GFP+ cells in the paranigral region and the interfascicular nucleus.
Since Sox6 and Otx2 have complementary expression (Panman et al., 2014), we analyzed Otx2 expression in the cumulative and the post-mitotic analyses. In Sox6-Cre, Th-2A-Flpo, RC-Frepe brains (Figure S6C), the OTX2 signal was low/residual in SNc neurons with no apparent difference between GFP+ and mCherry+ cells. In contrast, in the VTA, several neurons co-expressed GFP and OTX2. In Sox6-FSF-Cre, Th-2A-Flpo, RC-Frepe brains, most OTX2+ cells in the VTA were mCherry+ (Figure S6D). These results suggest that Sox6+ progenitors can contribute to Otx2+ neurons. It is possible that the sporadic labeled cells observed in the lateral progenitor domain gave rise to some Otx2+ VTA neurons. Alternatively, medial Sox6+ progenitors could contribute to Otx2+ neurons, indicating some developmental flexibility.
The OTX2 staining also confirmed our analysis in Figure S4 that distinguished VTA from dSNc/SNpl. We found that many mCherry cells in the VTA expressed Otx2 robustly, but mCherry populations in the dSNc/SNpl showed weak/residual OTX2 signal. We observed similar results with Lpl expression, which was robust in ventromedial VTA neurons, but low/residual in dSNc/SNpl populations (not shown). This confirms that the dSNc/SNpl populations are distinct from mCherry+ cells in the VTA. To investigate whether this difference manifests early in development, we examined OTX2 expression in E14.5 embryos. Rostral sections had fewer OTX2+,mCherry+ cells in contrast to caudal sections, where many are observed (Figure S6E), suggesting that rostral mCherry+ cohorts contribute to the dSNc and SNpl, while caudal mCherry cohorts contribute to the OTX2+ ventromedial VTA.
Finally, we investigated how the Sox6+ cohort was represented in Aldh1a1+, Calb1+, and Vglut2+ populations in adult Sox6-Cre, Th-2A-Flpo, RC-Frepe brains. In the SNc, most ALDH1A1+ neurons co-expressed GFP, 85% ± 3% (Figure 5F). Conversely, in the VTA, 77.4% ± 1.4% of ALDH1A1+ neurons co-expressed mCherry (not shown). These data strongly suggest that the Aldh1a1+ population has predominantly Sox6+ history in the SNc but not in the VTA. CB+ neurons, however, appear to lack Sox6 expression in both areas. The majority of CB+ neurons were mCherry+ in the SNc (80% ± 3%) (Figure 5G) and in the VTA (84.7 ± 5%; data not shown). Interestingly, in the VTA, the proportion of GFP+,CB+ neurons was slightly higher in the cumulative versus the postmitotic analysis, confirming our finding of more GFP+ neurons in the VTA of Sox6-Cre brains (Figure 5E). Overall, aside from the small GFP+,CB+ population, most Calb1+ neurons never expressed Sox6; therefore, they must derive from Sox6− progenitors. Next, we analyzed Vglut2, a marker that appears almost complementary to Sox6 (Figure S4). We performed in situ hybridization and found that 12% ± 2% of the TH+ SNc neurons expressed Vglut2, consistent with previous reports (Kouwenhoven et al., 2020). Expectedly, Vglut2+,TH+ neurons were heavily biased toward the mCherry+ cohort in the SNpl (85.6% ± 3% of Vglut2+ DA neurons were mCherry+; Figure 5H) and in the VTA (92.2 ± 8%, not shown). In the SNc, 48.5% ± 6% of the Vglut2+ DA neurons co-expressed mCherry (not shown). These numbers suggest that Vglut2+ neurons, especially in the SNpl and VTA, lack sufficient Sox6 expression to drive recombination throughout their history. In conclusion, dSNc/SNpl neurons that have never experienced significant levels of Sox6 in their history must predominantly originate from embryonic Sox6− progenitors, a key finding of our work. The Sox6 dorsoventral division of the SNc must be, at least in part, encoded in the FP progenitors, strongly suggesting a dual developmental origin of the SNc.
Sox6+ FP progenitors are biased toward a vSNc fate
Our cumulative Sox6 analysis showed that Sox6− dSNc neurons (mCherry+ and Calb1+) must predominantly originate from Sox6− progenitors. Sox6+ neurons (GFP+ and Aldh1a1+) in the vSNc, however, either derived from Sox6+ progenitors or acquired Sox6 expression postmitotically. Since cumulative fate maps cannot distinguish whether Sox6 expression occurred in progenitors, post-mitotic neurons, or both, we used our PRISM (progenitor restricted intersectional fate mapping) approach to obtain a bona fide Sox6 fate map (Poulin et al., 2020b). PRISM elements include a Cre driver (in this case, Sox6-Cre) a Nestin-driven Flpo recombinase Nes-LSL-Flpo (NSF), and a Flpo-dependent reporter, RC-LacZ, encoding for β-galactosidase (βGal) (Figure 6A). Flpo expression depends on Cre and Nestin; thus, the reporter is restricted to progenitors expressing Sox6, and it excludes labeling of DA neurons expressing Sox6 postmitotically (Lendahl et al., 1990).
Figure 6. Sox6 progenitor fate map.
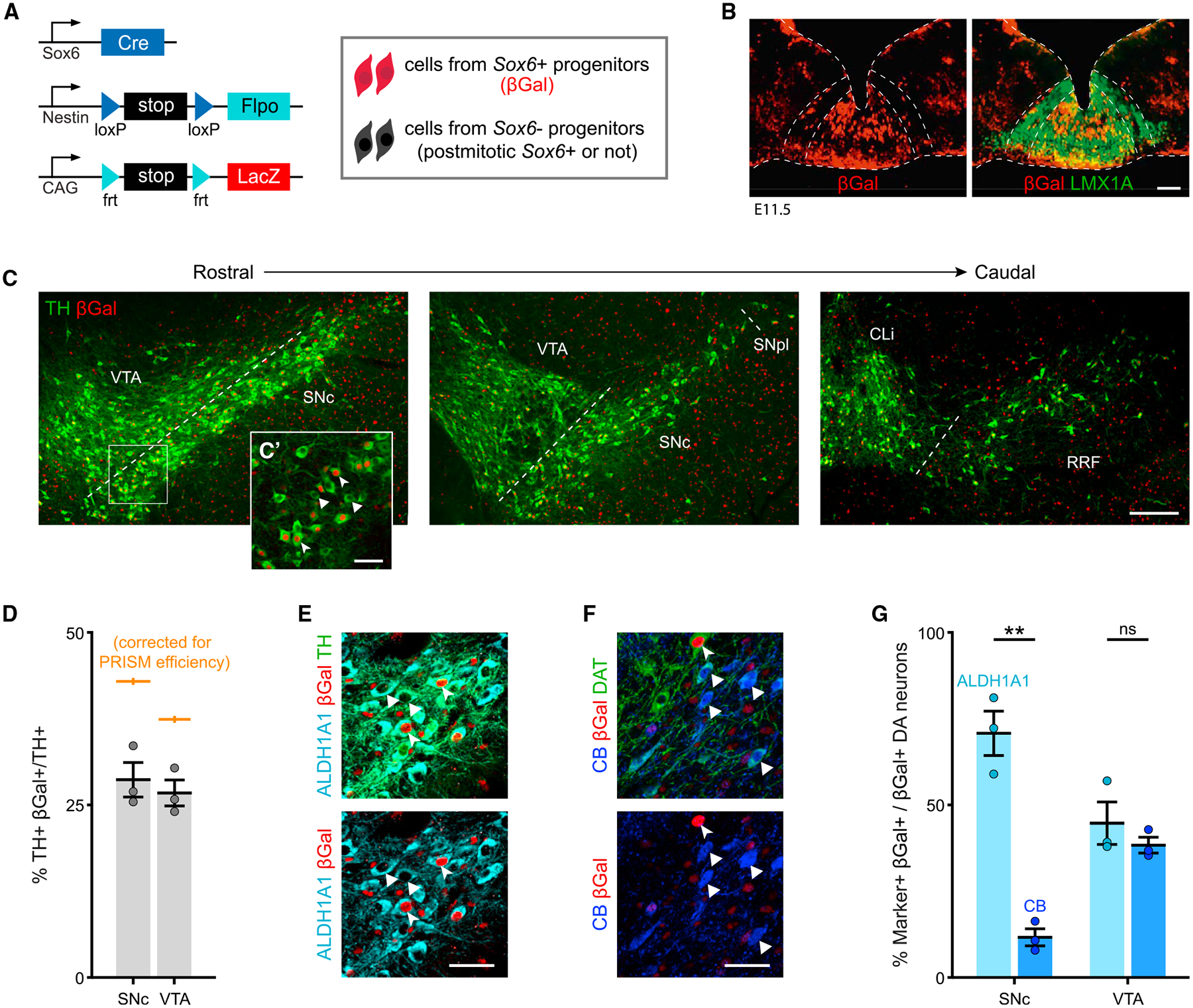
(A) Schematic of PRISM alleles.
(B) Sox6-Cre, Nestin-LSL-Flpo, RC-LacZ (from here on Sox6 PRISM) at E11.5. βGal+ progenitors (Sox6+) are located medially in the Lmx1a+ floor plate.
(C) Sox6 PRISM adult brains show TH+,βGal+ neurons in the SNc and VTA (n = 8).
(C’) High magnification of SNc TH+,βGal+ cells (arrowheads) and TH+,βGal− cells (triangles).
(D) Average TH+,βGal+/TH+ neurons in Sox6 PRISM adult brains, also corrected by PRISM efficiency; p = 0.1.
(E) High magnification of Sox6 PRISM SNc neurons co-expressing TH, ALDH1A1, and βGal.
(F) Sox6 PRISM SNc cells co-expressing dopamine transporter (DAT), CALBINDIN-D28k (CB), and βGal. Most CB+,DAT+ cells are βGal− (triangles). For reference, arrowhead shows a βGal+,DAT+,CB− cell.
(G) Percentage of βGal+ cells that express ALDH1A1 or CB in the SNc and VTA of Sox6 PRISM adult brains (p values: SNc p = 0.008, VTAp = 0.5).
Scale bars: (C) 200 μm; all other scale bars, 50 μm. Error bars are SEMs. (B and D–G) n = 3.
First, we validated the progenitor specificity of PRISM using Vglut2-Cre, since Vglut2 is expressed in nascent post-mitotic DA neurons (Kouwenhoven et al., 2020) (Figure S7A). As expected, few to no TH+ cells were labeled with the reporter in adult brains (Figure S7B), confirming that PRISM does not label post-mitotic neurons expressing the Cre driver. Second, we determined the maximum recombination potential of PRISM specifically in DA neurons, using EIIa as Cre driver, since EIIa is active at the one/two-cell stage (Figure S7C). We calculated an efficiency of 66.7% ± 2% in the SNc and 71.5% ± 0.01% in the VTA (Figure S7D), in line with the overall PRISM efficiency (Poulin et al., 2020b). This efficiency level provides a more complete picture of descendants, compared to genetically induced fate mapping strategies.
To confirm progenitor labeling, we analyzed Sox6-Cre, Nestin-LSL-Flpo, RC-LacZ embryos at E11.5. Sox6+ progenitors (βGal+) were largely localized medially in the embryonic Lmx1a+ domain (Figure 6B). Since not every medial progenitor was labeled, we compared Sox6-Cre, Nestin-LSL-Flpo, Ai65F (Flpo reporter encoding for tdTomato) to Sox6-Cre, Ai9 (Cre reporter encoding for tdTomato) at E12.5, the peak of DA neurogenesis. In brains of both genotypes, tdTomato+ cells were biased toward the medial Lmx1a+ domain (Figures S7E and S7F). Furthermore, the proportion of tdTomato+,LMX1A+/LMX1A+ cells in the PRISM embryo was 77% of the Sox6-Cre, Ai9 embryo (Figure S7F), roughly consistent with the overall PRISM efficiency (Poulin et al., 2020b) and the efficiency in DA neurons (Figure S7D).
PRISM provides a progenitor-restricted fate map in the adult brain. In adult Sox6 PRISM brains, we observed βGal+ DA neurons in the SNc, as well as the VTA (Figure 6C). We also observed βGal+,TH− cells in the rostral linear nucleus (not shown), characterized by low TH immunoreactivity. In the SNc, 28.7% ± 2.5% of the TH+ neurons originated from Sox6+ progenitors (42.9%, adjusted by PRISM efficiency; Figure 6D). To rule out possible recombination inefficiency due to a rox-flanked destabilized EGFP cassette in the Nestin-LSL-Flpo construct, we deleted the cassette using Dre-deleter mice to reduce the distance between the loxP sites. The adult fate map with a Dre-deleted Nes-LSL-Flpo showed a result similar to that of the undeleted construct (Figure S7G). Notwithstanding our several efforts to account for efficiency, we interpret our data cautiously to include the possibility of some amount of temporal delay due to sequential recombination. Therefore, we conclude that at minimum, ~40% of the SNc is derived from Sox6+ progenitors, although this could be an underestimation.
Next, we sought to determine the propensity of Sox6 progenitors to contribute to Sox6, Aldh1a1, and Calb1 populations. In the SNc ~92% of TH+,βGal+ fate-mapped neurons maintained SOX6 expression, whereas in the VTA, a substantial proportion of TH+,βGal+ were SOX6− (not shown), further supporting that some VTA neurons originate from Sox6+ progenitors but downregulate Sox6. Moreover, in Sox6-Cre PRISM brains, we found that 70.7% ± 6% of the βGal+ neurons co-expressed ALDH1A1+ (Figures 6E and 6G), and 11.6% ± 2.4% of the βgal+ neurons co-expressed CB (Figures 6F and 6G); these CB+ neurons tended to be located more medially in the SNc. We concluded that Sox6+ progenitors have a higher propensity to generate Sox6+ and Aldh1a1+ neurons than Calb1+ neurons.
To corroborate our findings, we generated a Sox6-CreERT2 knockin line (Figure S7H). Although this approach suffers from substantial mosaicism, among other limitations, it is a well-established technique to temporally restrict the activation of the reporter (Joyner and Zervas, 2006). In Sox6-CreERT2, Ai9 E11.5 brains, tdTomato+ progenitors localized mainly to the medial domain of the embryonic FP, as expected (Figure S7I). We followed the fate of the reporter-labeled cells in P0 brains and observed tdTomato+,TH+ neurons in the SNc, but also in the VTA/interfascicular region (Figures S7J and S7K). Our results indicate that Sox6+ progenitors contribute to a significant number of SNc neurons, but also to the VTA.
DISCUSSION
Despite emerging data on DA neuron heterogeneity, little is known about its developmental basis. This study demonstrates a dual embryonic origin of the SNc that results in neuronal cohorts with different molecular characteristics, projections, functions, PD pathways, and vulnerability (Figure 7).
Figure 7. Distribution of Sox6+ and Sox6− neurons, their projections, and developmental origin.
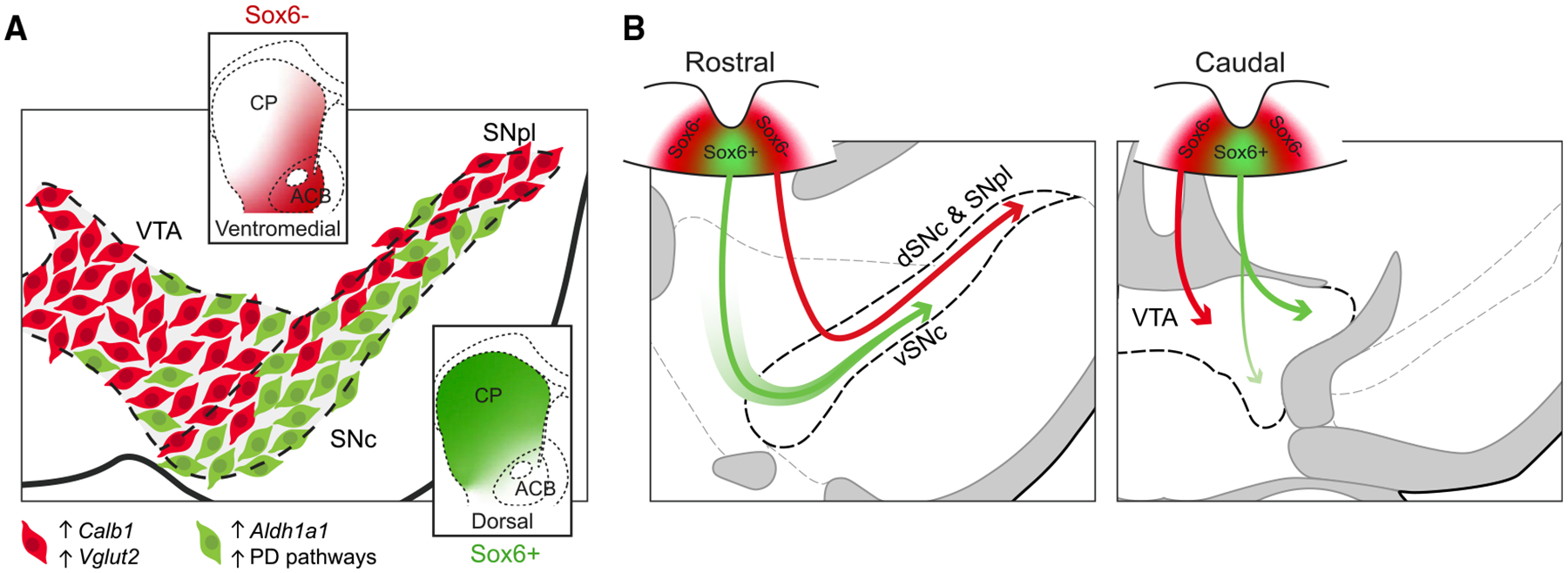
(A) In an adult midbrain section, Sox6+ DA neurons (green) ventrally biased in the SNc and in the parabrachial VTA are enriched in Aldh1a1 as well as in PD pathways and electron transport pathways. These neurons project to the dorsal striatum. Sox6− DA neurons (red) are enriched in Calb1 and/or Vglut2, and project to the medial, ventral, and tail of the striatum.
(B) Model depicting main contribution of Sox6 progenitors to the SNc: Sox6+ and Sox6− progenitors give rise to vSNc and dSNc/SNpl neurons, respectively (left), indicating a dual developmental origin of the SNc. Some neurons in the vSNc can also acquire Sox6 expression postmitotically (green gradient surrounding arrow). Main contribution of Sox6 progenitors to the VTA: in more caudal regions of the embryo, Sox6− progenitors predominantly give rise to ventromedial VTA neurons (right), and RRF neurons (not shown). However, Sox6+ progenitors give rise to neurons localized in the parabrachial region, some of which maintain Sox6 expression (solid green arrow), while others localized in the paranigral region downregulate Sox6 (gradient green arrow). In these more caudal regions, smaller contributions of Sox6+ and Sox6− can be found in the SNc and RRF.
The recent identification of putative DA subtypes (Poulin et al., 2020a) motivated us to examine whether preferential loss of one of them underpins the anatomically based observations on selective vulnerability in PD (Damier et al., 1999; Kordower et al., 2013; Mendez et al., 2005; Yamada et al., 1990). Here, we show the presence of a ventrally located SOX6+,ALDH1A1+ population in human control brains, and that these neurons are selectively diminished in post-mortem PD brains; in contrast, CB+ cells were resilient. Our findings are consistent with previous reports using ALDH1A1 and CALB1 as markers (Liu et al., 2014; Yamada et al., 1990), as well as recent studies depicting ventral enrichment of SOX6+ neurons in the human SNc (Kamath et al., 2021; Monzón-Sandoval et al., 2020). In mice, we found that Sox6+ neurons displayed large transcriptomic differences from Sox6− neurons that are likely to underpin their different properties (Figure 7A). Sox6+ neurons are enriched in pathways for ATP synthesis and therefore may carry a higher bioenergetic burden than Sox6− neurons (Surmeier et al., 2017). Increased ATP synthesis is likely required to support their continual activity during locomotion, but over time this may lead to oxidative stress in this population. Interestingly, immune-related gene sets are also enriched ventrally, including those for TLR signaling and major histocompatibility complex class II (MHC class II) pathways, both of which recently have been implicated in PD pathogenesis (Cebrián et al., 2014; Dzamko et al., 2017). Our study also showed that dorsal Sox6− cells were enriched for Calb1 and Vglut2 expression. Vglut2 has been recently identified as a marker of resilient DA neurons in toxin models, likely conferring resistance by way of Bdnf/TrkB signaling or by the facilitation of cytosolic DA clearance into vesicles (Shen et al., 2018; Steinkellner et al., 2018). Vglut2+ DA neurons have also been observed in primate SNc (Root et al., 2016). Calb1, however, could confer resiliency based on its calcium buffering properties (Inoue et al., 2019; Surmeier et al., 2017). Overall, our study reveals molecular distinctions between Sox6+ and Sox6− SNc neurons, some of which likely account for their selective vulnerability.
Sox6− dorsal and Sox6+ ventral SNc neurons showed distinctive projection patterns (Figure 7A). Extending our previous work (Poulin et al., 2018), here, we show that Sox6+ neurons densely innervate the DS; however, with a subtractive strategy, not used in Poulin et al., we reveal that Sox6− axons sparsely innervate the DS and instead project more densely to the medial and ventral striatum, including potentially part of the lateral ACB. An inverted projection pattern was previously reported in rodents and primates (Fallon and Moore, 1978; Gerfen, 1985; Haber et al., 2000), but it has not been observed consistently across all studies (Matsuda et al., 2009; Prensa and Parent, 2001); therefore, it has not been depicted in recent models of SNc projections to the CP (Björklund and Dunnett, 2007; Vogt Weisenhorn et al., 2016). Potential overlap of Sox6+ and Sox6− axonal arbors as shown here and in a recent study (Simmons et al., 2019), together with the diverging criteria used to define the dorsoventral division of the SNc and the partially intermingled nature of these neurons, all may be reasons for these inconsistent findings. Importantly, these anatomical findings can explain the patterns of striatal innervation observed in PD brains (Kordower et al., 2013); fibers projecting to the medial striatum are likely preserved because they originate from relatively spared Sox6− neurons in the dSNc, most of which are also Calb1+.
The Sox6− cohort also extends into the SNpl. Using single-cell transcriptomic datasets, we were able to identify Sox6− cells, Vglut2+ and Calb1+, and we localize these to the SNpl. Consistent with previous genetically and anatomically defined projection maps (Kim et al., 2014; Menegas et al., 2018; Poulin et al., 2018), we find substantial innervation of the TS. The SNpl-to-TS projection has recently been shown to have distinctive properties and encode threat prediction (Menegas et al., 2018). Watabe-Uchida and Uchida (2018) have postulated that this population may be considered a separate and orthogonal DA axis to the traditional reward-encoding axis. Our findings that these SNpl neurons are predominantly part of the developmentally encoded Sox6− cohort support the notion that these neurons have distinct functional properties.
Recent studies have revealed that the activity of some SNc neurons is time locked to movement initiation and acceleration, thereby rejecting the dogma that slow variations in firing (tens of seconds to minutes) in these same neurons bias animals toward or away from movement (Howe and Dombeck, 2016). A study measuring DA axon calcium transients in the CP suggested that axons encoding locomotion and reward signals were distinct; transients time locked to locomotion were prominent in the dorsal CP, but reward-locked transients were prominent in the ventral CP (Howe and Dombeck, 2016). Here, we extend these findings by demonstrating that Sox6+ cohorts encompass a population with calcium transients time locked to acceleration. Although further work is required to understand whether the Sox6+ cohort is homogeneous in its locomotion response, together with our data showing selective degeneration of SOX6+,ALDH1A1+ neurons in PD and the projections of the Sox6+ cohort to the dorsal CP, we propose a role for the Sox6+ cohort in motor control. This conclusion is consistent with a recent study showing that ablation of ~80% of Aldh1a1+ neurons in the SNc results in motor learning and walking speed deficits (Wu et al., 2019). Contrary to axons of Sox6+ neurons, those of Sox6− neurons densely project to the ventromedial striatum and the ACB, and respond to reward stimuli.
Toward understanding the basis of SNc neuron diversity, we provide evidence that the SNc has a dual embryonic origin (Figure 7B). From our multi-pronged lineage analysis, we find that the dSNc/SNpl is predominantly derived from Sox6− progenitors. These progenitors are enriched in the lateral FP. More caudally, these lateral FP progenitors give rise to Otx2+ neurons in the ventromedial VTA (Figures 7 and S6), consistent with the model of Panman et al. (2014). In contrast, Sox6+ progenitors enriched in the medial FP give rise to neurons in the SNc, with a greater propensity for Aldh1a1+ compared to Calb1+ fates. However, some of the cells in the vSNc originate from Sox6− progenitors but upregulate Sox6 post-mitotically. This conclusion finds support in the following observations: (1) some transient double-reporter-positive cells are observed at embryonic stages in Sox6-Cre, Th-2A-Flpo-based experiments (Figure S6B), and (2) even after accounting for PRISM efficiency, our progenitor restricted fate map does not label the entire vSNc. Nonetheless, because of potential temporal delay due to sequential recombination in PRISM fate maps, in conjunction with the slightly earlier birth of the SNc (Bye et al., 2012), it is difficult to exclude the possibility that a greater fraction of the vSNc was derived from Sox6+ progenitors. Overall, however, we provide evidence that SNc neuron fate is, at least in part, encoded in FP progenitors. Subsequent molecular refinements, coupled with differential migrations, and axon guidance mechanisms, then culminate in the elaborate and diversified midbrain DA system seen in the adult (Bodea et al., 2014; Brignani et al., 2020; Chabrat et al., 2017; La Manno et al., 2016; Prasad and Pasterkamp, 2009; Tiklová et al., 2019; Vaswani et al., 2019).
Sox6+ progenitors also contribute to DA neurons in the VTA, many of which are located in the parabrachial region and which continue to maintain Sox6 expression. However, some descendants were also observed in more ventromedial VTA regions, although some of these downregulate Sox6 during differentiation. The latter finding is supported by the observation that several GFP+ cells of the VTA are OTX2+ (Figure S6), and it suggests that all DA progenitors may not be completely fixed in their lineage. This would be in accordance with recent postulates on cell fate determination in the CNS, in which neurons converge on stable states through a process of gradient descent, using “configurational codes” that are somewhat plastic and evolve during development (Mayer et al., 2018). An alternative explanation could be that some low-level Sox6-expressing cells in the lateral progenitor domain, superseded the threshold of recombination and contributed to the VTA fate map.
Understanding DA neuron diversity is a key step toward clarifying the myriad functions of midbrain DA neurons. Leveraging the power of intersectional/subtractive genetic methods and Sox6 as an anchor, we provide a developmental perspective into how this diversity may be generated in the early embryo, and in doing so, we provide clarification to adult taxonomic schemes. Further work will be required to define molecules that may induce Sox6 in the FP, as well as define the functional role and direct targets of Sox6, toward the diversification of DA neurons.
Limitations of the study
In addition to the technical limitations that have been discussed throughout the Results section, we highlight the following: human—post-mortem human studies should be followed up with anterior-posterior stereological analyses; mouse—whether Sox6− populations are truly negative or express low levels that are below either the recombination threshold or the sensitivity of single-cell RNA-seq, remains to be determined. Despite this caveat, we were able to show that these genetically defined populations are distinct in several different ways. With respect to our fate-mapping experiments, we deduced that dorsal SNc/SNpl neurons originated from Sox6− progenitors; future experiments should aim to directly label lateral FP progenitors and determine their fate.
STAR★METHODS
RESOURCE AVAILABILITY
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Rajeshwar Awatramani (r-awatramani@northwestern.edu).
Materials availability
Mouse lines generated in this study will be shared upon request upon completion of Material Transfer Agreement due to institutional policy and will be deposited to a mouse repository (e.g., MMRRC).
Data and code availability
Bulk RNaseq data have been deposited at Gene Expression Omnibus and are publicly available as of the date of publication.
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the Lead Contact upon request.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Patient information
Tissue samples, clinical and neuropathology data, were supplied by the Parkinson’s UK Brain Bank funded by Parkinson’s UK, a charity registered in England, in Wales (258197) and Scotland (SC037554). In total, fifteen PD patients between 67–92 years of Braak stages V-VI were evaluated. Cases were age-matched to controls (details in Table S1). All post-mortem analyses were approved by the Ethical Research Committee of the Centre Hospitalier Universitaire de Québec (#2016–2564).
Mice
All animals used in this study were maintained and cared following protocols approved by the Northwestern Animal Care and Use Committee. Cre and Flpo mouse lines were maintained heterozygous by breeding to wild-type C57BL6 mice. Reporter lines were maintained homozygous. The Sox6-FSF-Cre and Sox6-FSF-CreERT2 lines were generated at Northwestern University by the Transgenic and Targeted Mutagenesis Laboratory as previously described (Poulin et al., 2018). To obtain Sox6-FSF-CreERT2, we replaced the Cre cassette with a CreERT2 cassette (Poulin et al., 2018). We crossed Sox6-FSF-Cre and Sox6-FSF-CreERT2 to CAG-Flpe to obtain Sox6-Cre and Sox6-CreERT2 lines, respectively.
For postmitotic or cumulative analyses, we crossed Sox6-FSF-Cre or Sox6-Cre to Th-2A-Flpo (Poulin et al., 2018), RC-Frepe (JAX 029486) and harvested brains at P56 and P21-P30, respectively. Both males and females were used for all adult brain analyses. As reported in Poulin et al. (2018), Th-2A-Flpo has > 99% recombination efficiency in DA neurons. For projection and functional analyses, Sox6-FSF-Cre mice were crossed to Th-2A-Flpo mice, and offspring were used for viral injections at 3 to 5 months old. To obtain progenitor labeling at E11.5, Sox6-FSF-Cre animals were crossed to CAG-Flpe, RC-Frepe (Figure 5A). To obtain progenitor labeling at E12.5, Sox6-Cre animals were crossed to Ai9 (JAX 007909). For PRISM analysis, we used our newly generated Nes-LSL-Flpo (NSF) line (Poulin et al., 2020b), Ella-Cre (JAX 003724), Slc17a6-Cre (Vglut2) (JAX 028863), and Sox6-Cre, crossed to RC-FA (RC-LacZ), that was derived by crossing the intersectional reporter RC-Fela (Jensen et al., 2008) (gift from S. Dymecki) with the beta-actin Cre-deleter mouse (JAX 003376). To establish maximum efficiency of PRISM, Nes-LSL-Flpo was first crossed to EIIa-Cre. Offspring with a deletion of the loxP-STOP-loxP cassette (Nes-Flpo) was next crossed to RC-FA. This breeding strategy assured that loxP-STOP-loxP cassette preceding Flpo was excised in the germline. To permanently delete the rox-flanked destabilized eGFP cassette (d4GFP) in Nes-LSL-Flpo, this PRISM line was crossed to a Dre-deleter mice as previously described (Poulin et al., 2020b). Brains of adult males and females were harvested at P21 for all PRISM-related experiments mentioned above. For progenitor-anchored fate maps at E11.5 and E12.5, Nes-LSL-Flpo was crossed to Sox6-Cre with RC-FA and Ai65F (JAX 032864), respectively. For genetically induced fate map experiments, tamoxifen (Sigma Aldrich, prepared in corn oil) was administered by IP injection to pregnant dams of E9.5 Sox6-CreERT2, Ai9 (2 mg/40 g). For all embryonic time points, the morning of the day when a vaginal plug was detected was designated as E0.5.
METHOD DETAILS
Identification of anatomical borders in human tissue by Nissl staining
We referred to published work (Damier et al., 1999; Mai and Paxinos, 2012; Olszewski and Baxter, 1954) to define human substantia nigra (hSNc) anatomical borders and used Nissl staining to confirm the rostro-caudal level of the sections. Sections were deparaffinized by heated treatment (65°C), treated with citrisolv, (Decon laboratories Inc., #1601) rehydrated in ethanol gradients (70%, 50% and 25% for 2 min) and incubated in cresyl violet solution for 20 min followed by dehydration in ethanol gradients (25%, 50%, 70%, 95%, 100%) and citrisolv. Slides were coverslipped with DPX mounting medium (Electron Microscopy Sciences #13512) and observed under a Nikon Eclipse E800 bright field microscope.
Immunofluorescence in hSNc
Sections were deparaffinized followed by incubation in citrisolv, hydration steps with ethanol gradients (100%, 95%, 70%, and 50%) and quick rinse in PBS - 0.2% Tween. Sections were next incubated in a pre-heated antigen retrieval solution of 10mM citrate buffer (pH6.2) (Sigma-Aldrich, #W302600–1KG-K) for 30 min at 95°C and cooled for 1 hour at RT. Sections were then rinsed in wash buffer (PBS - 0.2% Tween - 5% BSA (BioShop, ALB001.500) - 2.5% Triton X (Sigma-Aldrich, T8787) and incubated in blocking solution (5% BSA, 2.5% Triton X, 10% donkey serum (Sigma-Aldrich, D9663) for 1 hour at RT. Finally, sections were incubated overnight at RT in blocking solution with the following primary antibodies: rabbit anti-SOX6 (Sigma-Aldrich, #HPA001923, 1:100), sheep anti-TH (Pel-Freez Biologicals, #P60101–0, 1:1000), mouse anti-ALDH1A1 (Sigma, #SAB5300519, 1:200), rabbit anti-CALBINDIN-D-28K (Millipore, #AB1778, 1:500). Subsequently, sections were rinsed three times in wash buffer. Secondary antibodies were diluted in blocking solution and incubated for 2 hours and 30 min at RT with Alexa Fluor 488 donkey anti-rabbit IgG (Life Technologies, #A21206, 1:500), Alexa Fluor 546 donkey anti-sheep IgG (Life Technologies, #A21098, 1:500) and Alexa Fluor 647 donkey anti-mouse IgG (Life Technologies, #A 31571, 1:500). Sections were then washed three times and incubated 7 min at RT in DAPI (Molecular probes by Life Technologies, #D3571), rinsed 3 times, washed for 5 min in ethanol 70% and incubated for 5 min at RT in autofluorescence eliminator reagent (Millipore, #2160) followed by three washes in ethanol 70% for 1 min. Slides were ultimately dried and coverslipped using fluoromount G (Electron microscopy, #17984–25). To ensure antibodies specificity, positive and negative controls were included. For SOX6 positive control, human tissue was obtained from cases with brain metastases which are known to be immune reactive for SOX6. Negative controls were performed by omitting the primary antibody. Images of human SNc (hSNc) were acquired on the slide scanner Zeiss Axio Scan.Z1 with Zen 2.3 lite software. High magnifications were obtained on Zeiss LSM 800 confocal laser microscope with Zen 2.1 software. Images were acquired by z series and reconstructed in Fiji software.
Immunofluorescence in mouse tissue
Embryonic brains were harvested and fixed in 1% or 4% PFA-PBS for an amount of time depending upon age, cryoprotected in 30% sucrose-PBS before OCT embedding and sectioning at 16–20 μm on a cryostat. Adult brains were perfused and fixed overnight with 4% PFA-PBS, cryoprotected in 30% sucrose-PBS solution, snap frozen in dry-ice and sectioned at 20–25 μm on a microtome. Sections were rinsed in PBS and blocked in 5% normal donkey serum in PBS 0.3% Triton X for 30 min at room temperature (overnight at 4°C for staining with goat anti-Aldh1a1), then incubated with primary antibodies diluted in blocking solution overnight at 4°C. Antigen retrieval: Tissue was post-fixed 5 min in 4% PFA-PBS, rinsed in PBS, treated with antigen unmasking solution (Vector Laboratories) and heated in a microwave for 5–10 minutes depending on the primary antibody, then blocked for 1 hour and incubated overnight at 4°C with primary antibodies diluted in the blocking solution. The following day, sections were rinsed in PBS and incubated with secondary antibodies at room temperature, followed by PBS rinse and mounting. Primary antibodies: rabbit and sheep anti-TH (Pel-Freez, 1:500 and 1:250, respectively), chicken anti-GFP (Abcam, 1:1500), rabbit anti-mCherry (Abcam, 1:1000), rat anti-mCherry (Abcam, 1:1000), mouse anti-CB (Abcam, 1:500), rabbit anti-OTX2 (Proteintech, 1:500), goat anti-ALDH1A1 (Abcam, 1:1000), rabbit anti-Lmx1A (Millipore, 1:1000, antigen retrieval), guinea pig anti-LMX1A (Gift from Dr. Yongchao C. Ma, 1:20,000, antigen retrieval), goat anti-βGal (Biogenesis, 1:1500), rabbit anti-RFP (LSBio, 1:1000, antigen retrieval), rat anti-DAT (Santa Cruz, 1:200). Secondary antibodies: Alexa 488, −555, and −647 (Molecular Probes) or Cy3 and Cy5 (Jackson Immuno Research), diluted 1:250 in blocking solution with DAPI (1mg/mL; Sigma).
In situ hybridization
RNAscope assay was performed on 4% PFA fixed brains (as explained above), using the fluorescent multiplex kit from Advanced Cell Diagnostic according to manual instructions for detection of Slc17a6, (Vglut2) (Mm-Slc17a6-C3; ACD #319171) Sst, (Mm-Sst-C3; ACD #404631) Ngb, (Mm-Ngb-C3; ACD #876171) Fxyd6, (Mm-Fxyd6-C3; ACD #430971). We combined this method with standard immunofluorescence for mCherry and TH as explained above, performing RNAscope first. 16 μm sections were obtained with a cryostat. Slides were coverslipped with ProLong Gold antifade reagent (Sigma). We determined Vglut2 positivity by counting the number of puncta surrounding a cell’s nucleus. All cells were observed to have basal expression of Vglut2. Any cell that had 10 or more distinct, clustered, puncta were considered Vglut2 positive. All images for this study were obtained on Olympus BX61VS slide scanner, confocal Nikon A1R-spectral microscope, Nikon W1 Dual Cam Spinning Disk Confocal at Northwestern University Center for Advanced Microscopy/Nikon Imaging Center, or Leica TCE SPE DMI4000 B (Dr. Dimitri Krainc, Northwestern University). Figures were prepared in Adobe Illustrator.
Isolation of dorsal and ventral SNc for transcriptome analysis
Sox6-FSF-Cre, Th-2A-Flpo, RC-Frepe adult brains (P56 and older) were used for bulk RNaseq analysis. Anesthetized mice were rapidly decapitated and the SNc was carefully dissected at ~1 mm coronal thickness using a brain matrix (Kent Scientific #RBMS-200C) and further trimmed under an epi-fluorescent microscope, avoiding other dopaminergic populations. Samples were processed with the Papain Dissociation System (Worthington Biochem #LK003150) following the manufacturer’s recommendations. However, we substituted the provided EBSS solution with ACSF buffer (200 mM Sucrose, 2.6 mM KCl, 10 mM MgCl2, 0.5 mM CaCl2, 26 mM NaHCO3, 1.27 mM NaH2PO4 and 10 mM Dextrose; equilibrated with 5% CO2/95% O2; pH 7.3). Tissue was dissociated in 20U papain per mL in ACSF, 1mM of L-cysteine, and 0.5mM EDTA. At the end, cells pellets were re-suspended in 400 μL of cold ACSF enriched with 1% B27, 1% BSA and DAPI, then FACS sorted. Roughly an equal number of mCherry+ (Sox6−) and GFP+ (Sox6+) cells were collected from 3 independent animals, and each sample was processed individually. Total RNA was prepared according to Ambion mirVana miRNA isolation kit (Life technology, AM1561).
RNA-sequencing and analysis
The stranded mRNaseq was conducted in the Northwestern University NUSeq Core Facility. Total RNA samples were checked for quality using RINs generated from Agilent Bioanalyzer 2100. RNA quantity was determined with Qubit fluorometer. The Illumina TruSeq Stranded mRNA Library Preparation Kit was used to prepare sequencing libraries of high-quality RNA samples (RIN > 7). The Kit procedure was performed without modifications. This procedure includes mRNA enrichment and fragmentation, cDNA synthesis, 3′ end adenylation, Illumina adaptor ligation, library PCR amplification and validation. Illumina NextSeq500 NGS Sequencer was used to sequence the libraries with the production of single-end, 75 bp reads. The quality of reads, in FASTQ format, was evaluated using FastQC (v0.11.7). Reads were trimmed to remove Illumina adapters from the 3′ ends using cutadapt (v1.14) (Martin, 2011). Trimmed reads were aligned to the Mus musculus genome (mm10/GRCm38.p6) using STAR (version 020201) (Dobin et al., 2013). Read counts for each gene were calculated using htseq-count (version 0.6.1p1) (Anders et al., 2015) in conjunction with a gene annotation file for mm10 obtained from Ensembl (http://useast.ensembl.org/index.html). Normalization and differential expression were calculated using DESeq2 (version 1.14.1) that employs the Wald test (Love et al., 2014). The cutoff for determining significantly differentially expressed genes (Table S2) was an FDR-adjusted p value less than 0.05 using the Benjamini-Hochberg method. A pathway analysis was performed using Metascape (http://metascape.org) to identify significant pathways among the significantly differently expressed genes.
Data visualization
Volcano plot: After aligning the sequences and calculating the number of reads obtained for each gene, we used DESeq2 (Version 3.10) (Love et al., 2014) to estimate fold-change (FC) difference and significance between GFP+ and mCherry+ populations. DESeq was applied to the normalized counts using negative binomial distribution. False discovery rate (FDR < 0.1) and −1.4 < FC > 1.4 were used as the parameters to select genes that have a significant expression change. GSEA: To analyze enrichment of gene sets between GFP+ and mCherry+ populations, we used GSEA software v4.0.2, freely available from the Broad Institute (https://www.gsea-msigdb.org/gsea/%3chttps://urldefense.proofpoint.com/v2/url?u=http-3A__software.broadinstitute.org_gsea_index.jsp&d=DwMFAw&c=yHlS04HhBraes5BQ9ueu5zKhE7rtNXt_d012z2PA6ws&r=q3PUTvWH-k7Xg3m5Xu4sgxcoX1GSmOsPCLxG3Gp_YWuadKi8GxI1hS07DWXW9OVf&m=DxuT1tLCsjQY122nzFQBXGrXiIEWgjUk267rtpz0REI&s=bENoDJQckIp8TSIOCehptZttSrNMD9kAuhGiAJgdyjQ&e=.) (Mootha et al., 2003; Subramanian et al., 2005). First, we converted genes in our mouse RNaseq dataset to human homologs using the Human and Mouse Homologs list available from The Jackson Laboratory website (http://www.informatics.jax.org/homology<https://urldefense.proofpoint.com/v2/url?u=http-3A__www.informatics.jax.org_homology&d=DwMFAw&c=yHlS04HhBraes5BQ9ueu5zKhE7rtNXt_d012z2PA6ws&r=q3PUTvWH-k7Xg3m5Xu4sgxcoX1GSmOsPCLxG3Gp_YWuadKi8GxI1hS07DWXW9OVf&m=DxuT1tLCsjQY122nzFQBXGrXiIEWgjUk267rtpz0REI&s=HZWtPgdN1VRlokcmHqekpXhS8nAhYslkvjbdefMOZLg&e=>). A total of 15828 genes were subsequently used for GSEA. Next, we compared GFP+ to mCherry+ populations using a curated collection of gene sets, “Canonical Pathways,” available from MSigDB (Liberzon et al., 2011; Subramanian et al., 2005). All default settings were used for the analysis, except for a minimum gene set size of 25 and use gene set for permutation type (recommended by GSEA for analyzing fewer than seven samples). Heatmap: we used pheatmap R package (version 1.0.12) to build a heatmap from reads counts. Log2FC threshold was set to 0.65, and alpha to 0.05. Genes were filtered by FDR < 0.05 to obtain the heatmap.
Projections in adult brains
The SNc of Sox6-FSF-Cre, Th-2A-Flpo adults (6–8 weeks old, n = 3) were injected with AAV5 -hSyn-CreOn/FlpOn-EYFP UNC AV8357 and AAV8 EF1α-CreOff/FlpOn-mCherry (gift from K. Deisseroth) viruses (Figures 4A, 4B, and S5A). For Figure S5D, viruses used were AAV5-EF1α-DIO-mCherry #AV4311B (abbreviated as AAV-CreOn-mCherry) and AAVdj-hSyn-CreOff/FlpOn-eYFP (gift from K. Deisseroth) (n = 3). Four weeks later, brains were perfused with 4% PFA-PBS and fixed overnight in 4% PFA-PBS. The following day, after PBS washes, brains were processed to prepare 25μm sections on a freezing microtome. Immunofluorescence for mCherry and GFP (eYFP) was performed on floating sections as described above. Epifluorescence images were acquired on an Olympus Slide Scanner VS120.
For the histograms of Figures 4E and 4F, sections for CPr and CPi of n = 3 mice above were used. A rectangle was drawn for each section in dorso-ventral and medio-lateral direction, as depicted in Figures 4E and 4F, left. These areas were divided into 25 bins and the % of total fluorescence for each mCherry and eYFP was calculated in each bin.
One-photon fiber photometry
For calcium fiber photometry, adult mice (6–8 weeks old) of Sox6-FSF-Cre, Th-2A-Flpo genotype were injected within the SNc with either AAV8 EF1α-CreOn/FlpOn-GCaMP6f or AAV8-EF1α-CreOff/FlpOn-GCaMP6f (gifts from K. Deisseroth), respectively (Figure 4G). n = 9 mice were used for Sox6+ recordings, and n = 10 for Sox6−. Mice were head-fixed over a cylindrical treadmill and allowed to run freely. Non-predicted water rewards were delivered through a waterspout, at random intervals. 200μm core optic fibers were placed into dorsal striatum for Sox6+ recordings (X ± 1.8, Y 0.5, Z 1.9 mm) and in the ventral striatum for Sox6− recordings (X ± 1.8, Y 0.5, Z 3.9 mm). A 470nm LED was used for illumination and emitted light was diverged onto the PMT using a 505-sp dichroic and a 540nm/50 filter. Fluorescence was recorded at 4000 Hz and binned at 100 Hz. Mice with no significant transients were excluded (none for Sox6+ and 8 for Sox6−. We confirmed through histology that in most mice few Sox6− axons were labeled, not enough to reach the detection threshold of fiber photometry). %ΔF/F was calculated as in Howe and Dombeck (2016) Triggered averages were calculated for reward delivery and accelerations exceeding 1ms−2 GCaMP expression and fiber placement were checked by immunofluorescence for GFP performed on floating sections as described above.
QUANTIFICATION AND STATISTICAL ANALYSIS
Human samples
For quantification, the hSNc was first identified and delineated on each slide using Nissl staining. Adjacent slides were used for staining where anatomical delimitations were reproduced. The shape of each region was delineated using Zen 2.3 lite software and both single and double-labeled neurons were counted. Student’s t tests were used between two means with a Welch correction when the homogeneity of variance could not be confirmed with Bartlett’s test. Two-way ANOVA followed by Tukey’s post hoc analyses was used when two categorical data such as “group,” “sex” or “levels of analyzed brains” were investigated. No significant statistical differences between sex were identified. Likelihood ratio analysis of contingency tables with Pearson’s method was used in the investigation of categorical data such as “sex.” P values under 0.05 were considered significant. All statistical analyses were performed using Prism (Graphpad). Data were expressed as mean ± SEM and each point depicted on the graphs represents one sample.
Mouse samples
We referred to the mouse brain Allen Reference Atlas (by Wiley) and Paxinos G. & Franklin K. B (second edition by Academic Press) to identify brain section levels and establish anatomical boundaries in conjunction with TH expression. Multi-channel images were processed in Fiji to draw boundaries and perform cell counts. All counted cells were DAPI+ and all DA neurons counted were TH+ or DAT+. Specifically, GFP+ and mCherry+ cells counted for Figures 2C–2E, 5E, S2B, S2C, and S2E, were also TH+. Cell counts on adult brains were performed on three brains per genotype and three to six matching sections across the rostro-caudal axis (Allen atlas levels B-2.88 to B-3.52 for SNc, VTA, SNpl and −3.88 for RRF). We quantified 3 sections for all experiments with CB and ALDH1A1 staining, and for PRISM efficiency in adult brains. For Sox6-FSF-Cre, Th-2A-Flpo, RC-Frepe analysis in Figure 2 and S2, we quantified 5 brains, 4 sections each. For in situ hybridization of Vglut2, we quantified 3 brains, 6 sections each. For quantifications at E12.5, we analyzed 3 sections per brain, 3 brains per genotype.
For the first approach to characterize spatial distribution of reporter-labeled cells, we performed a bisection of the SNc for dorsal-ventral analysis performed in ImageJ. First, a dorsal and a ventral SNc boundary was drawn. The length of each boundary was divided into 10 segments, marked by ticks on the boundary line. Starting from one end, a vertical line was drawn to join the first tick on the dorsal boundary to the first tick on the ventral boundary, the process was repeated for the subsequent 9 ticks. Finally, a bisector line was drawn by crossing each midpoint of the 10 vertical lines. mCherry+TH+ and GFP+TH+ cells were counted on each half. These data are not included in figures but are discussed in the results section.
For the second approach, to characterize spatial distribution of reporter-labeled cells, we plotted histograms of cell spatial distribution across the dorso-ventral axis of the SNc in Figures 2D, 2E, and S2C (example diagram Figures S2A–S2A”‘). xy coordinates for mCherry+TH+ and GFP+TH+ cells were obtained with ImageJ cell counter plugin. The angle and the dorso-ventral center of the SNc was obtained using a line of best fit given all SNc cells, and dorso-ventral coordinates were defined perpendicular to this line. Bins for the histograms are 15 μm wide. For Figures 2E and S2C mCherry and GFP cells were normalized separately by relative probability (sum of each histogram = 100%). The centroids for each population on each section were plotted on Figure 2E right. 4 rostro-caudal sections were analyzed for each n = 3 mice. In Figure S2C, histograms were calculated separately for each rostro-caudal section. In Figures 2D and 2E, all sections for each mouse were counted together. Shaded area is SEM. For Figure S7F, we obtained xy coordinates for all LMX1A+ and tdTomato+ cells in 3 brains with ImageJ cell counter plugin, 3 sections each. A vertical line was drawn at the midline, and distance to the midline calculated for each cell. For left 2 plots, a histogram (5.5 μm bin) was obtained for each strategy. For the right plot, we calculated the % of tdTomato+ cells at each distance from the midline for each strategy.
For genetically induced fate mapping analysis (Figures S7H–S7K), we used six animals and at least ten TH+ midbrain sections were counted per brain. All values were expressed as mean ± SEM. Student’s t test was used for statistical comparisons and p values lower than 0.05 were considered significant. Analysis was performed in MATLAB.
Supplementary Material
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Chicken Polyclonal anti-GFP | Abcam | Cat# ab13970; RRID: AB_300798 |
| Rat Monoclonal anti-mCherry | Thermo Fisher | Cat# M11217; RRID: AB_2536611 |
| Goat Polyclonal anti-βgal | Bio-Rad | Cat# 4600–1409; RRID: AB_2307350 |
| Mouse monoclonal anti-CALBINDIN-D-28K | Sigma | Cat# C9848; RRID: AB_476894 |
| Rabbit Polyclonal anti-OTX2 | Proteintech | Cat# 13497–1-AP; RRID: AB_2157176 |
| Goat Polyclonal anti-Aldh1a1 | R&D Systems | Cat# AF5869; RRID: AB_2044597 |
| Rabbit Polyclonal anti-Sox6 | Sigma | Cat# HPA001923; RRID: AB_1080065) |
| Guinea pig Polyclonal anti-Lmx1a | Gift from Yongchao C. Ma | N/A |
| Rabbit Polyclonal anti-Lmx1a | Millipore | Cat# AB10533; RRID: AB_10805970 |
| Rabbit Polyclonal anti-CALBINDIN-D-28K | Millipore | Cat# AB1778; RRID: AB_2068336 |
| Mouse Monoclonal anti-ALDH1A1 | Sigma | Cat# SAB5300519 |
| Rat Polyclonal anti-DAT | Santa Cruz Biotech | Cat# sc-32258; RRID: AB_627400 |
| Mouse Monoclonal anti-RFP | Abcam | Cat# ab125244; RRID: AB_10973556 |
| Sheep Polyclonal anti-TH | Pel-Freez | Cat# P60101–0; RRID: AB_461070 |
| Chemicals, peptides, and recombinant proteins | ||
| Tamoxifen | Sigma | T5648 |
| Experimental models: cell lines | ||
| PRX-B6 (C57BL/6N) (mouse ES cells) | The Jackson Laboratory | 012448; RRID: IMSR_JAX:012448 |
| Experimental models: mouse strains | ||
| Sox6-FSF-Cre | Poulin et al. 2018 | N/A |
| Sox6-Cre | This paper | N/A |
| Sox6-CreERT2 | This paper | N/A |
| Th-2A-Flpo | Poulin et al. 2018 | N/A |
| Mouse: Tg(EIIa-cre)C5379Lmgd | The Jackson Laboratory | 003724; RRID: IMSR_JAX:003724 |
| Mouse: Slc17a6tm2(cre)Lowl | The Jackson Laboratory | 016963; RRID: IMSR_JAX:016963 |
| NSF | (Poulin et al., 2020b) | N/A |
| B6;129S6-Gt(ROSA)26Sortm8(CAG-mCherry,-EGFP) Dym/J | The Jackson Laboratory | 029486; RRID: IMSR_JAX:029486 |
| RC-Fela | A gift from S. Dymecki (Jensen et al. 2008) | N/A |
| Mouse: B6.Cg-Gt(ROSA)26Sortm65.2(CAG-tdTomato) Hze/J | The Jackson Laboratory | 032864; RRID: IMSR_JAX:032864 |
| Mouse: B6;129S6-Gt(ROSA)26Sortm9(CAG-tdTomato) Hze/J | The Jackson Laboratory | 007905; RRID: IMSR_JAX:007905 |
| Mouse: B6;129-Tg(CAG-dre)1Afst/Mmucd | MMRRC | 032246-UCD; RRID: MMRRC_032246-UCD |
| Oligonucleotides | ||
| Primers for Cre strains Cre-F: GCAGAACCTGAAGATGTTCGC | This paper | N/A |
| Primers for Cre strain Cre-R: ACACCAGAGACGGAAATCCATC | This paper | N/A |
| Primers for NSF F: CACCAAGACCAAGACCCTGT | This paper | N/A |
| Primers for NSF R: CCTTCAGCAGCTGGTACTCC | This paper | N/A |
| Primers for RC-Fela, RC-Frepe, RC-Ai9, RC-Ai65F -F: TGCAATACCTTTCTGGGAGTTC | This paper | N/A |
| Primers for RC-Fela, RC-Frepe, RC-Ai9, RC-Ai65F -R: AGCGGGAGAAATGGATATGAAG | This paper | N/A |
| Primers for RC-Fela, RC-Frepe, RC-Ai9, RC-Ai65F -R: TACCGTAAGTTATGTAACGCGG | This paper | N/A |
| Primers for Th-2A-Flpo F- TAAGACCCTGCTGATGGTTGG | This paper | N/A |
| Primers for Th-2A-Flpo R- CATAGGGCATTCCTGTGGTTTG | This paper | N/A |
| Primers for Th-2A-Flpo R- GCTTCACTGAGTCTCTGGCATC | This paper | N/A |
| Recombinant DNA | ||
| AAV5-hSyn-CreOn/FlpOn-EYFP | UNC | # AV8357 |
| AAV8EF1α-CreOff/FlpOn-mCherry | gift from K. Deisseroth | #2466 |
| AAV5-EF1α-DIO-mCherry | UNC | #AV4311B |
| AAVdj-hSyn-CreOff/FlpOn-eYFP | gift from K. Deisseroth | #987 |
| AAV8-EF1α-CreOn/FlpOn-GCaMP6f | gift from K. Deisseroth | #2383 |
| AAV8-EF1α-CreOff/FlpOn-GCaMP6f | gift from K. Deisseroth | #2385 |
| Deposited data | ||
| RNaseq of GFP and mCherry cells of Sox6-FSF-Cre, Th-2A-Flpo, RC- Frepe SNc | This Paper | GEO: GSE185480 |
| Software and algorithms | ||
| VS-ASW-S6 | Olympus | https://www.olympus-lifescience.com/en/microscopes/virtual/vs120/ |
| cellSens | Olympus | https://www.olympus-lifescience.com/en/software/cellsens/ |
| Zen 2.3 and 3.3 | Zeiss | https://www.zeiss.com/microscopy/us/products/microscope-software/zen.html |
| MATLAB | MathWorks | https://www.mathworks.com/products/matlab.html |
| RStudio | RStudio | https://www.rstudio.com/ |
| FastQC | Babraham Bioinformatics | https://www.bioinformatics.babraham.ac.uk/projects/fastqc/ |
| STAR | Dobin et al., 2013 | https://github.com/alexdobin/STAR |
| DESeq2 | Bioconductor |
https://bioconductor.org/packages/release/bioc/html/DESeq2.html https://doi.org/10.18129/B9.bioc.DESeq2. |
| cutadapt | cutadapt |
https://cutadapt.readthedocs.io/en/stable/ DOI: 10.14806 |
| pheatmap | CRAN | https://cran.r-project.org/web/packages/pheatmap/index.html |
| GSEA | UCSD | https://www.gsea-msigdb.org/gsea/index.jsp |
| ImageJ | NIH | https://imagej.nih.gov/ij/ |
Highlights.
The SNc derives from Sox6+ and Sox6− progenitors, indicating a dual origin
Sox6+ and Sox6− DA neuron cohorts have distinctive projections to the striatum
Sox6+ DA neurons are enriched in pathways and genes that may underpin vulnerability
Sox6+,Aldh1a1+ neurons are selectively vulnerable in PD
ACKNOWLEDGMENTS
We thank Michael Wegner (guinea pig anti-Sox6 antibody), Maria Grazia Spillantini (human anatomical boundaries), Vasileios Papakis (confocal imaging), and Matthew Schipma (RNA-seq analysis). This work was supported by NIH grants R01-NS119690, R01-NS096240, R01-MH110555, R21-NS092034-01A1, P50 DA044121-01A1, and 1 F31 NS122481-01A1.
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental information can be found online at https://doi.org/10.1016/j.celrep.2021.109975.
DECLARATION OF INTERESTS
The authors declare no competing interests.
REFERENCES
- Anders S, Pyl PT, and Huber W (2015). HTSeq–a Python framework to work with high-throughput sequencing data. Bioinformatics 31, 166–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersson E, Tryggvason U, Deng Q, Friling S, Alekseenko Z, Robert B, Perlmann T, and Ericson J (2006). Identification of intrinsic determinants of midbrain dopamine neurons. Cell 124, 393–405. [DOI] [PubMed] [Google Scholar]
- Arendt D, Bertucci P, Achim K, and Musser JM (2019). Evolution of neuronal types and families. Curr. Opin Neurobiol 56, 144–152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Björklund A, and Dunnett SB (2007). Dopamine neuron systems in the brain: an update. Trends Neurosci. 30, 194–202. [DOI] [PubMed] [Google Scholar]
- Blaess S, Bodea GO, Kabanova A, Chanet S, Mugniery E, Derouiche A, Stephen D, and Joyner AL (2011). Temporal-spatial changes in Sonic Hedgehog expression and signaling reveal different potentials of ventral mesencephalic progenitors to populate distinct ventral midbrain nuclei. Neural Dev. 6, 29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodea GO, Spille J-H, Abe P, Andersson AS, Acker-Palmer A, Stumm R, Kubitscheck U, and Blaess S (2014). Reelin and CXCL12 regulate distinct migratory behaviors during the development of the dopaminergic system. Development 141, 661–673. [DOI] [PubMed] [Google Scholar]
- Brignani S, Raj DDA, Schmidt ERE, Düdükcü Ö, Adolfs Y, De Ruiter AA, Rybiczka-Tesulov M, Verhagen MG, van der Meer C, Broekhoven MH, et al. (2020). Remotely Produced and Axon-Derived Netrin-1 Instructs GABAergic Neuron Migration and Dopaminergic Substantia Nigra Development. Neuron 107, 684–702.e9. [DOI] [PubMed] [Google Scholar]
- Brown A, Machan JT, Hayes L, and Zervas M (2011). Molecular organization and timing of Wnt1 expression define cohorts of midbrain dopamine neuron progenitors in vivo. J. Comp. Neurol 519, 2978–3000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bye CR, Thompson LH, and Parish CL (2012). Birth dating of midbrain dopamine neurons identifies A9 enriched tissue for transplantation into parkinsonian mice. Exp. Neurol 236, 58–68. [DOI] [PubMed] [Google Scholar]
- Cebrián C, Zucca FA, Mauri P, Steinbeck JA, Studer L, Scherzer CR, Kanter E, Budhu S, Mandelbaum J, Vonsattel JP, et al. (2014). MHC-I expression renders catecholaminergic neurons susceptible to T-cell-mediated degeneration. Nat. Commun 5, 3633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chabrat A, Brisson G, Doucet-Beaupré H, Salesse C, Schaan Profes M, Dovonou A, Akitegetse C, Charest J, Lemstra S, Côté D, et al. (2017). Transcriptional repression of Plxnc1 by Lmx1a and Lmx1b directs topographic dopaminergic circuit formation. Nat. Commun 8, 933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- da Silva JA, Tecuapetla F, Paixão V, and Costa RM (2018). Dopamine neuron activity before action initiation gates and invigorates future movements. Nature 554, 244–248. [DOI] [PubMed] [Google Scholar]
- Damier P, Hirsch EC, Agid Y, and Graybiel AM (1999). The substantia nigra of the human brain. I. Nigrosomes and the nigral matrix, a compartmental organization based on calbindin D(28K) immunohistochemistry. Brain 122, 1421–1436. [DOI] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, and Gingeras TR (2013). STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodson PD, Dreyer JK, Jennings KA, Syed ECJ, Wade-Martins R, Cragg SJ, Bolam JP, and Magill PJ (2016). Representation of spontaneous movement by dopaminergic neurons is cell-type selective and disrupted in parkinsonism. Proc. Natl. Acad. Sci. USA 113, E2180–E2188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dzamko N, Gysbers A, Perera G, Bahar A, Shankar A, Gao J, Fu Y, and Halliday GM (2017). Toll-like receptor 2 is increased in neurons in Parkinson’s disease brain and may contribute to alpha-synuclein pathology. Acta Neuropathol. 133, 303–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans RC, Zhu M, and Khaliq ZM (2017). Dopamine Inhibition Differentially Controls Excitability of Substantia Nigra Dopamine Neuron Subpopulations through T-Type Calcium Channels. J. Neurosci 37, 3704–3720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fallon JH, and Moore RY (1978). Catecholamine innervation of the basal forebrain. IV. Topography of the dopamine projection to the basal forebrain and neostriatum. J. Comp. Neurol 180, 545–580. [DOI] [PubMed] [Google Scholar]
- Farassat N, Costa KM, Stojanovic S, Albert S, Kovacheva L, Shin J, Egger R, Somayaji M, Duvarci S, Schneider G, and Roeper J (2019). In vivo functional diversity of midbrain dopamine neurons within identified axonal projections. eLife 8, e48408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiorenzano A, Sozzi E, Parmar M, and Storm P (2021). Dopamine Neuron Diversity: Recent Advances and Current Challenges in Human Stem Cell Models and Single Cell Sequencing. Cells 10, 1366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerfen CR (1985). The neostriatal mosaic. I. Compartmental organization of projections from the striatum to the substantia nigra in the rat. J. Comp. Neurol 236, 454–476. [DOI] [PubMed] [Google Scholar]
- Giguère N, Burke Nanni S, and Trudeau L-E (2018). On Cell Loss and Selective Vulnerability of Neuronal Populations in Parkinson’s Disease. Front. Neurol 9, 455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haber SN, Fudge JL, and McFarland NR (2000). Striatonigrostriatal pathways in primates form an ascending spiral from the shell to the dorsolateral striatum. J. Neurosci 20, 2369–2382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hook PW, McClymont SA, Cannon GH, Law WD, Morton AJ, Goff LA, and McCallion AS (2018). Single-Cell RNA-Seq of Mouse Dopaminergic Neurons Informs Candidate Gene Selection for Sporadic Parkinson Disease. Am. J. Hum. Genet 102, 427–446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howe MW, and Dombeck DA (2016). Rapid signalling in distinct dopaminergic axons during locomotion and reward. Nature 535, 505–510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue KI, Miyachi S, Nishi K, Okado H, Nagai Y, Minamimoto T, Nambu A, and Takada M (2019). Recruitment of calbindin into nigral dopamine neurons protects against MPTP-induced parkinsonism. Mov. Disord 34, 200–209. [DOI] [PubMed] [Google Scholar]
- Jensen P, Farago AF, Awatramani RB, Scott MM, Deneris ES, and Dymecki SM (2008). Redefining the serotonergic system by genetic lineage. Nat. Neurosci 11, 417–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joksimovic M, Anderegg A, Roy A, Campochiaro L, Yun B, Kittappa R, McKay R, and Awatramani R (2009). Spatiotemporally separable Shh domains in the midbrain define distinct dopaminergic progenitor pools. Proc. Natl. Acad. Sci. USA 106, 19185–19190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joyner AL, and Zervas M (2006). Genetic inducible fate mapping in mouse: establishing genetic lineages and defining genetic neuroanatomy in the nervous system. Dev. Dyn 235, 2376–2385. [DOI] [PubMed] [Google Scholar]
- Kamath T, Abdulraouf A, Burris S, Gazestani V, Nadaf N, Vanderburg C, and Macosko EZ (2021). A molecular census of midbrain dopaminergic neurons in Parkinson’s disease. bioRxiv. 10.1101/2021.06.16.448661. [DOI] [Google Scholar]
- Kim HF, Ghazizadeh A, and Hikosaka O (2014). Separate groups of dopamine neurons innervate caudate head and tail encoding flexible and stable value memories. Front. Neuroanat 8, 120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kordower JH, Olanow CW, Dodiya HB, Chu Y, Beach TG, Adler CH, Halliday GM, and Bartus RT (2013). Disease duration and the integrity of the nigrostriatal system in Parkinson’s disease. Brain 136, 2419–2431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouwenhoven WM, Fortin G, Penttinen A-M, Florence C, Delignat-Lavaud B, Bourque M-J, Trimbuch T, Luppi MP, Salvail-Lacoste A, Legault P, et al. (2020). VGluT2 Expression in Dopamine Neurons Contributes to Postlesional Striatal Reinnervation. J. Neurosci 40, 8262–8275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer DJ, Risso D, Kosillo P, Ngai J, and Bateup HS (2018). Combinatorial Expression of Grp and Neurod6 Defines Dopamine Neuron Populations with Distinct Projection Patterns and Disease Vulnerability. eNeuro 5, ENEURO.0152–18.2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Manno G, Gyllborg D, Codeluppi S, Nishimura K, Salto C, Zeisel A, Borm LE, Stott SRW, Toledo EM, Villaescusa JC, et al. (2016). Molecular Diversity of Midbrain Development in Mouse, Human, and Stem Cells. Cell 167, 566–580.e19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lendahl U, Zimmerman LB, and McKay RDG (1990). CNS stem cells express a new class of intermediate filament protein. Cell 60, 585–595. [DOI] [PubMed] [Google Scholar]
- Lerner TN, Shilyansky C, Davidson TJ, Evans KE, Beier KT, Zalocusky KA, Crow AK, Malenka RC, Luo L, Tomer R, and Deisseroth K (2015). Intact-Brain Analyses Reveal Distinct Information Carried by SNc Dopamine Subcircuits. Cell 162, 635–647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liberzon A, Subramanian A, Pinchback R, Thorvaldsdóttir H, Tamayo P, and Mesirov JP (2011). Molecular signatures database (MSigDB) 3.0. Bioinformatics 27, 1739–1740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu G, Yu J, Ding J, Xie C, Sun L, Rudenko I, Zheng W, Sastry N, Luo J, Rudow G, et al. (2014). Aldehyde dehydrogenase 1 defines and protects a nigrostriatal dopaminergic neuron subpopulation. J. Clin. Invest 124, 3032–3046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love MI, Huber W, and Anders S (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mai JK and Paxinos G, eds. (2012). The Human Nervous System, Third Edition (Elsevier; ), p. xi. [Google Scholar]
- Martin M (2011). Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet. J 17, 10–12. [Google Scholar]
- Matsuda W, Furuta T, Nakamura KC, Hioki H, Fujiyama F, Arai R, and Kaneko T (2009). Single nigrostriatal dopaminergic neurons form widely spread and highly dense axonal arborizations in the neostriatum. J. Neurosci 29, 444–453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto M, and Hikosaka O (2009). Two types of dopamine neuron distinctly convey positive and negative motivational signals. Nature 459, 837–841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer C, Hafemeister C, Bandler RC, Machold R, Batista Brito R, Jaglin X, Allaway K, Butler A, Fishell G, and Satija R (2018). Developmental diversification of cortical inhibitory interneurons. Nature 555, 457–462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendez I, Sanchez-Pernaute R, Cooper O, Viñuela A, Ferrari D, Björklund L, Dagher A, and Isacson O (2005). Cell type analysis of functional fetal dopamine cell suspension transplants in the striatum and substantia nigra of patients with Parkinson’s disease. Brain 128, 1498–1510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menegas W, Akiti K, Amo R, Uchida N, and Watabe-Uchida M (2018). Dopamine neurons projecting to the posterior striatum reinforce avoidance of threatening stimuli. Nat. Neurosci 21, 1421–1430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metzakopian E, Lin W, Salmon-Divon M, Dvinge H, Andersson E, Ericson J, Perlmann T, Whitsett JA, Bertone P, and Ang S-L (2012). Genome-wide characterization of Foxa2 targets reveals upregulation of floor plate genes and repression of ventrolateral genes in midbrain dopaminergic progenitors. Development 139, 2625–2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monzón-Sandoval J, Poggiolini I, Ilmer T, Wade-Martins R, Webber C, and Parkkinen L (2020). Human-Specific Transcriptome of Ventral and Dorsal Midbrain Dopamine Neurons. Ann. Neurol 87, 853–868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mootha VK, Lindgren CM, Eriksson K-F, Subramanian A, Sihag S, Lehar J, Puigserver P, Carlsson E, Ridderstråle M, Laurila E, et al. (2003). PGC-1α-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat. Genet 34, 267–273. [DOI] [PubMed] [Google Scholar]
- Nalls MA, Blauwendraat C, Vallerga CL, Heilbron K, Bandres-Ciga S, Chang D, Tan M, Kia DA, Noyce AJ, Xue A, et al. ; 23andMe Research Team; System Genomics of Parkinson’s Disease Consortium; International Parkinson’s Disease Genomics Consortium (2019). Identification of novel risk loci, causal insights, and heritable risk for Parkinson’s disease: a meta-analysis of genome-wide association studies. Lancet Neurol. 18, 1091–1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nouri P, Götz S, Rauser B, Irmler M, Peng C, Trümbach D, Kempny C, Lechermeier C, Bryniok A, Dlugos A, et al. (2020). Dose-Dependent and Subset-Specific Regulation of Midbrain Dopaminergic Neuron Differentiation by LEF1-Mediated WNT1/b-Catenin Signaling. Front. Cell Dev. Biol 8, 988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olszewski J, and Baxter D (1954). Cytoarchitecture of the human brainstem. By Jerzy Olszewski and Donald Baxter. Published and distributed in North America for S. Karger by J. B. Lippincott Company, Philadelphia and Montreal. 1954. 199 pages. Price $16.00 (Reviewed by Gerhardt von Bonin). J. Comp. Neurol 101, 825. [Google Scholar]
- Panman L, Papathanou M, Laguna A, Oosterveen T, Volakakis N, Acampora D, Kurtsdotter I, Yoshitake T, Kehr J, Joodmardi E, et al. (2014). Sox6 and Otx2 control the specification of substantia nigra and ventral tegmental area dopamine neurons. Cell Rep. 8, 1018–1025. [DOI] [PubMed] [Google Scholar]
- Poulin J-F, Zou J, Drouin-Ouellet J, Kim K-YA, Cicchetti F, and Awatramani RB (2014). Defining midbrain dopaminergic neuron diversity by single-cell gene expression profiling. Cell Rep. 9, 930–943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poulin J-F, Tasic B, Hjerling-Leffler J, Trimarchi JM, and Awatramani R (2016). Disentangling neural cell diversity using single-cell transcriptomics. Nat. Neurosci 19, 1131–1141. [DOI] [PubMed] [Google Scholar]
- Poulin J-F, Caronia G, Hofer C, Cui Q, Helm B, Ramakrishnan C, Chan CS, Dombeck DA, Deisseroth K, and Awatramani R (2018). Mapping projections of molecularly defined dopamine neuron subtypes using intersectional genetic approaches. Nat. Neurosci 21, 1260–1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poulin J-F, Gaertner Z, Moreno-Ramos OA, and Awatramani R (2020a). Classification of Midbrain Dopamine Neurons Using Single-Cell Gene Expression Profiling Approaches. Trends Neurosci. 43, 155–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poulin J-F, Luppi MP, Hofer C, Caronia G, Hsu P-K, Chan CS, and Awatramani R (2020b). PRISM: A Progenitor-Restricted Intersectional Fate Mapping Approach Redefines Forebrain Lineages. Dev. Cell 53, 740–753.e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prakash N, Brodski C, Naserke T, Puelles E, Gogoi R, Hall A, Panhuysen M, Echevarria D, Sussel L, Weisenhorn DMV, et al. (2006). A Wnt1-regulated genetic network controls the identity and fate of midbrain-dopaminergic progenitors in vivo. Development 133, 89–98. [DOI] [PubMed] [Google Scholar]
- Prasad AA, and Pasterkamp RJ (2009). Axon guidance in the dopamine system. In Development and Engineering of Dopamine Neurons, Pasterkamp J, Smidt MP, and Burbach JPH, eds. (Springer; ), pp. 91–100. [DOI] [PubMed] [Google Scholar]
- Prensa L, and Parent A (2001). The nigrostriatal pathway in the rat: a single-axon study of the relationship between dorsal and ventral tier nigral neurons and the striosome/matrix striatal compartments. J. Neurosci 21, 7247–7260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Root DH, Wang H-L, Liu B, Barker DJ, Mód L, Szocsics P, Silva AC, Maglóczky Z, and Morales M (2016). Glutamate neurons are intermixed with midbrain dopamine neurons in nonhuman primates and humans. Sci. Rep 6, 30615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saunders A, Macosko EZ, Wysoker A, Goldman M, Krienen FM, de Rivera H, Bien E, Baum M, Bortolin L, Wang S, et al. (2018). Molecular Diversity and Specializations among the Cells of the Adult Mouse Brain. Cell 174, 1015–1030.e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen H, Marino RAM, McDevitt RA, Bi G-H, Chen K, Madeo G, Lee P-T, Liang Y, De Biase LM, Su T-P, et al. (2018). Genetic deletion of vesicular glutamate transporter in dopamine neurons increases vulnerability to MPTP-induced neurotoxicity in mice. Proc. Natl. Acad. Sci. USA 115, E11532–E11541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmons SC, Wheeler K, and Mazei-Robison MS (2019). Determination of circuit-specific morphological adaptations in ventral tegmental area dopamine neurons by chronic morphine. Mol. Brain 12, 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinkellner T, Zell V, Farino ZJ, Sonders MS, Villeneuve M, Freyberg RJ, Przedborski S, Lu W, Freyberg Z, and Hnasko TS (2018). Role for VGLUT2 in selective vulnerability of midbrain dopamine neurons. J. Clin. Invest 128, 774–788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stott SRW, and Barker RA (2014). Time course of dopamine neuron loss and glial response in the 6-OHDA striatal mouse model of Parkinson’s disease. Eur. J. Neurosci 39, 1042–1056. [DOI] [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, and Mesirov JP (2005). Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 102, 15545–15550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surmeier DJ (2018). Determinants of dopaminergic neuron loss in Parkinson’s disease. FEBS J. 285, 3657–3668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surmeier DJ, Obeso JA, and Halliday GM (2017). Selective neuronal vulnerability in Parkinson disease. Nat. Rev. Neurosci 18, 101–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiklová K, Björklund ÅKK, Lahti L, Fiorenzano A, Nolbrant S, Gillberg L, Volakakis N, Yokota C, Hilscher MM, Hauling T, et al. (2019). Single-cell RNA sequencing reveals midbrain dopamine neuron diversity emerging during mouse brain development. Nat. Commun 10, 581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaswani AR, Weykopf B, Hagemann C, Fried H-U, Brüstle O, and Blaess S (2019). Correct setup of the substantia nigra requires Reelin-mediated fast, laterally-directed migration of dopaminergic neurons. eLife 8, e41623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogt Weisenhorn DM, Giesert F, and Wurst W (2016). Diversity matters - heterogeneity of dopaminergic neurons in the ventral mesencephalon and its relation to Parkinson’s Disease. J. Neurochem 139 (Suppl 1), 8–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watabe-Uchida M, and Uchida N (2018). Multiple Dopamine Systems: Weal and Woe of Dopamine. Cold Spring Harb. Symp. Quant. Biol 83, 83–95. [DOI] [PubMed] [Google Scholar]
- Wu J, Kung J, Dong J, Chang L, Xie C, Habib A, Hawes S, Yang N, Chen V, Liu Z, et al. (2019). Distinct Connectivity and Functionality of Aldehyde Dehydrogenase 1a1-Positive Nigrostriatal Dopaminergic Neurons in Motor Learning. Cell Rep. 28, 1167–1181.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada T, McGeer PL, Baimbridge KG, and McGeer EG (1990). Relative sparing in Parkinson’s disease of substantia nigra dopamine neurons containing calbindin-D28K. Brain Res. 526, 303–307. [DOI] [PubMed] [Google Scholar]
- Yamaguchi T, Ehara A, Nakadate K, and Ueda S (2018). Tyrosine hydroxylase afferents to the interstitial nucleus of the posterior limb of the anterior commissure are neurochemically distinct from those projecting to neighboring nuclei. J. Chem. Neuroanat 90, 98–107. [DOI] [PubMed] [Google Scholar]
- Yan CH, Levesque M, Claxton S, Johnson RL, and Ang S-L (2011). Lmx1a and lmx1b function cooperatively to regulate proliferation, specification, and differentiation of midbrain dopaminergic progenitors. J. Neurosci 31, 12413–12425. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Bulk RNaseq data have been deposited at Gene Expression Omnibus and are publicly available as of the date of publication.
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the Lead Contact upon request.


