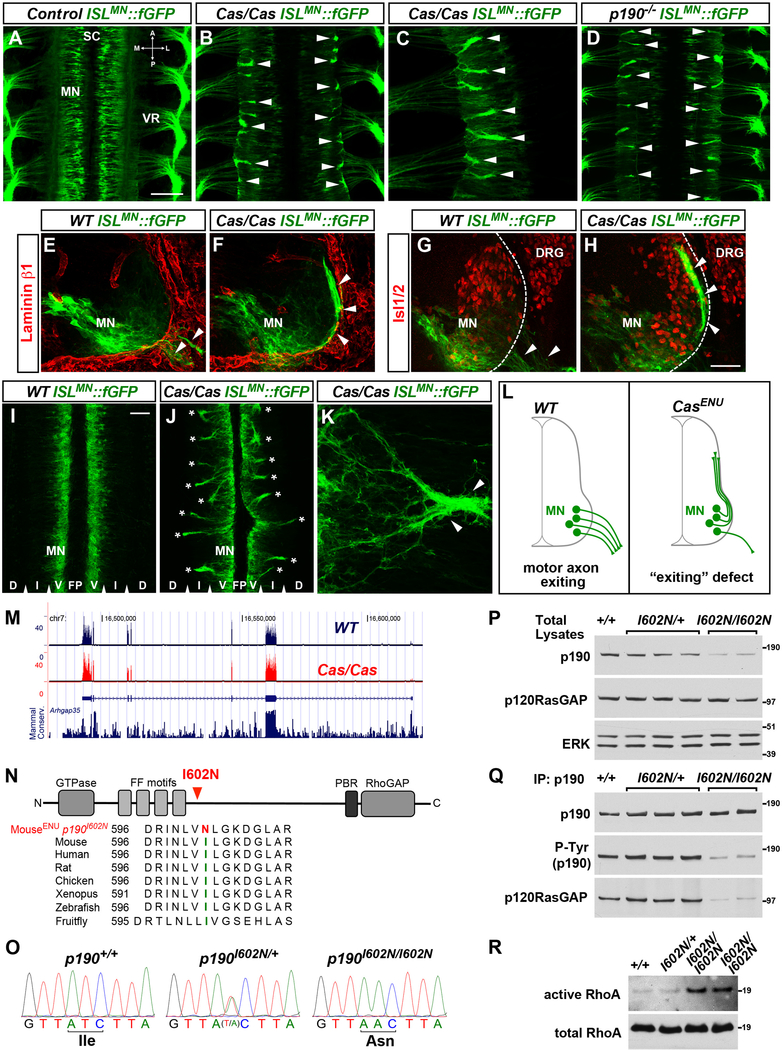
Figure 1. Cassin ENU Mutants Display Defects in Motor Axon Exit from the Spinal Cord.
(A–D) Whole mounts of ISLMN::fGFP+ mouse embryos (E12.5, dorsal views) show motor neuron cell bodies (MN) in the spinal cord (SC) and their axons exiting through the ventral roots (VR) in wild-type (A). Aberrant bundles of motor axons that fail to leave the spinal cord are visible in Cassin (B and C) and p190 mutants (D) (arrowheads). A, anterior; P, posterior; M, medial; L, lateral.
(E–H) ISLMN::fGFP+ motor neuron cell bodies (MN) and axons (arrowheads) in E11.5 transverse sections. Laminin-β1 (E and F) marks the spinal cord basal lamina (dashed line in G and H). Isl1/2 labels motor neurons and dorsal root ganglia (DRG). Motor axons select motor exit points in wild-type embryos (arrowheads in E and G) but form ectopic dorsally projecting bundles in Cassin mutants (arrowheads in F and H). ISLMN::fGFP signal is higher in medially positioned motor neurons.
(I–K) E12 spinal cord “open-book.” Bilateral columns of motor neuron somata (MN) separated by floor plate (FP), but no axons, are present in wild-types (I), whereas numerous intraspinal axon bundles spanning the intermediate (I) through dorsal (D) regions are detected in Cassin mutants (asterisks in J). (K) Ectopic intraspinal bundle (arrowheads) formed by convergence of misrouted axons in spinal open-book of Cassin embryo.
(L) Schematic of motor axon “exiting” phenotype of Cassin mutants.
(M) RNA-seq reads from spinal cord and adjacent tissues of E12.5 wild-type (blue trace) and Cassin (red trace) embryos aligned at the Arhgap35 locus (p190RhoGAP). Mammalian conservation track is reported. Exon coverage and levels of p190 mRNA are normal in mutants. RNA-seq did not detect global changes in gene expression or abnormal splicing in Cassin mutants.
(N) Schematic of p190RhoGAP and amino acid sequence alignment around isoleucine 602 (green) showing high evolutionary conservation and ENU-induced mutation I602N (red). p190 consists of a GTPase-like N-terminal domain competent for GTP binding, a “middle domain” with multiple protein-protein interaction motifs, and a C-terminal catalytic GAP domain.
(O) Genomic DNA sequencing identifies homozygous T > A substitution in p190 exon 1 of Cassin embryos, leading to I602N mutation.
(P) Decreased levels of p190RhoGAP in total lysates of E13.5 spinal cords of p190I602N/I602N (Cassin) mutant embryos compared to wild-type (+/+) and heterozygous (p190I602N/+) littermates. Levels of p120RasGAP, a known interactor of p190, are unchanged. ERK is a loading control.
(Q) Equal amounts of p190 were immunoprecipitated (IP) from spinal cord lysates used in (P). The levels of tyrosine-phosphorylated (P-Tyr) p190 and co-immunoprecipitated p120RasGAP detected by western blotting are reduced in p190I602N/I602N mutants.
(R) ncrease in GTP-bound RhoA (active form) pulled down from lysates of E13.5 p190I602N/I602N spinal cords compared to wild-type and heterozygous littermates (10.7- ± 2.2-fold increase p190I602N/I602N [n = 3] versus controls [n = 5]; mean ± SEM, Mann-Whitney U test p < 0.05).
Scale bars in (A), (B), and (D), 200 μm; in (C), 100 μm, in (F) and (G), 100 μm, in (H), 20 μm, and in (I)–(L), 50 μm. See also Figure S1.