FIG 1.
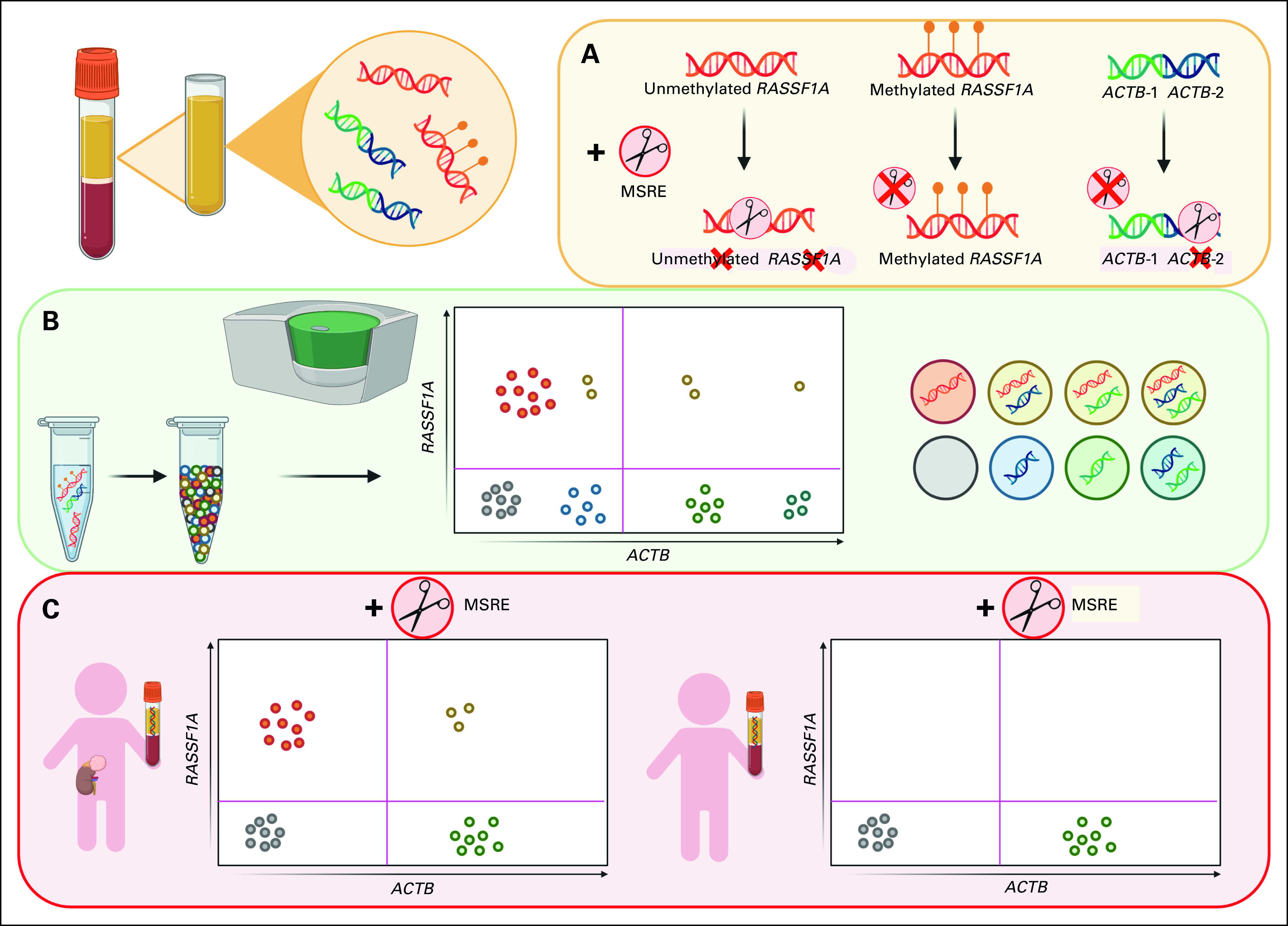
Concept of quantifying methylated RASSF1A using MSRE and ddPCR. (A) An MSRE incubation of a cfDNA sample results in the digestion of unmethylated RASSF1A, whereas methylated RASSF1A remains intact. Two amplicons of ACTB are added, and ACTB-1 is unaffected by the MSRE, whereas ACTB-2 is digested by the MSRE, as a control for MSRE performance. Every sample is subjected to two different ddPCR reactions, (B) one without the MSRE and (C) the other with the MSRE. ACTB-2 primers and probe are added in a lower concentration, resulting in a lower amplitude to discriminate between the ACTB-1 and ACTB-2 clusters. (C) Only in cfDNA from patients with circulating tumor DNA present, RASSF1A will be detected after digestion with the MSRE, as the absence of RASSF1A methylation will result in RASSF1A digestion, preventing the detection of this unmethylated RASSF1A allele by ddPCR. cfDNA, cell-free DNA; ddPCR, droplet digital polymerase chain reaction; MSRE, methylation-sensitive restriction enzymes.
