Figure 1: REV7 is a HORMA protein and is regulated through stable structural rearrangement.
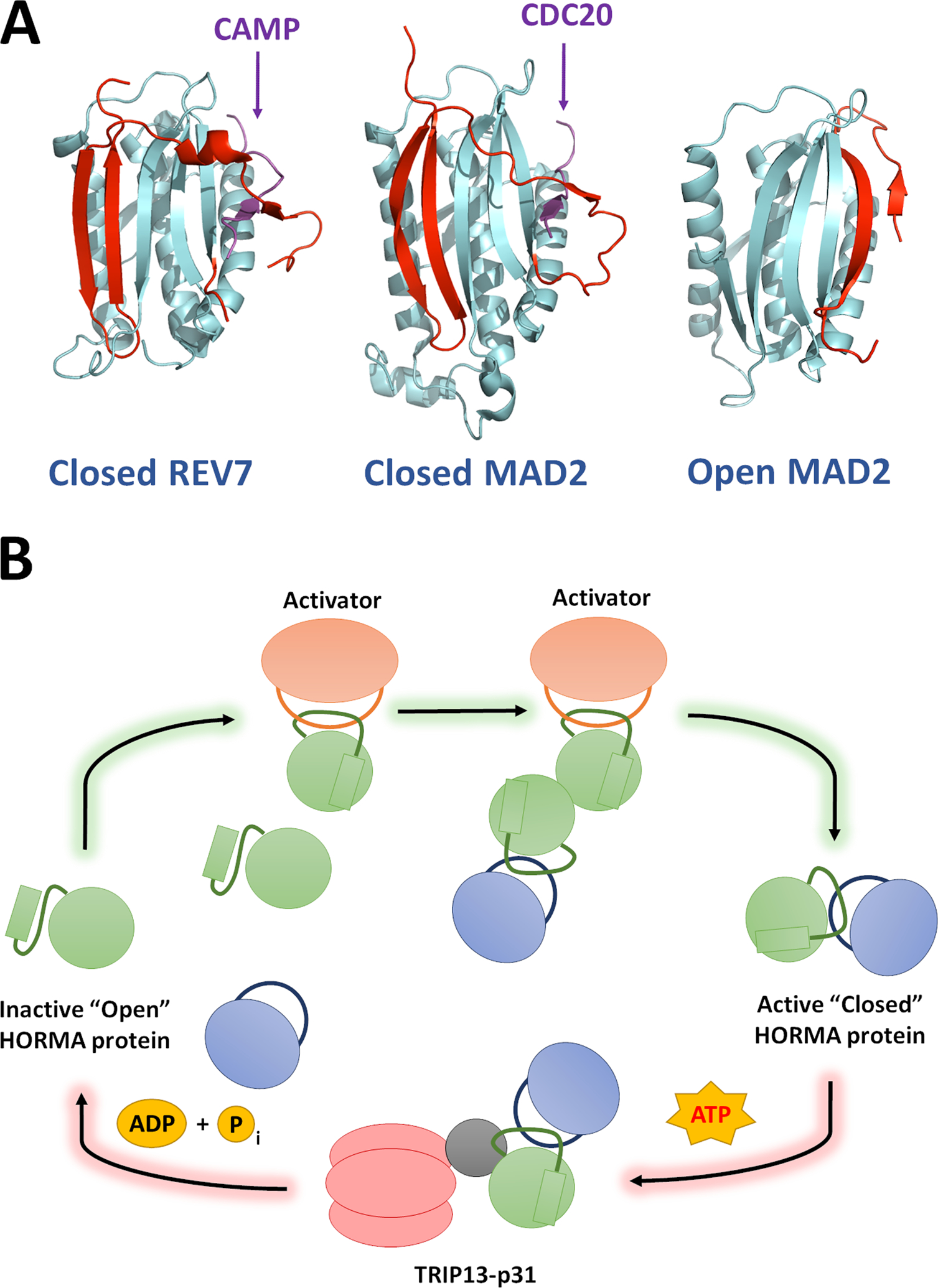
(A) Structures of REV7 and MAD2 in their closed, active forms (PDB ID: 5XPT and 4AEZ, respectively) and MAD2 in the open conformation. The seatbelt region of each protein is highlighted in red. REV7 and MAD2 show high structural similarity and bind to their cognate seatbelt binding partners via an identical mechanism. Small seatbelt-binding fragments of CAMP and CDC20 (Purple) are depicted in these representative structures. All known HORMA seatbelt binding partners associate though the same mechanism. (B) The “templating” model for HORMA protein activation: HORMA protein activation proceeds through a chain reaction mechanism, beginning when the first HORMA molecules are closed through their interaction with a dedicated activation partner. HORMA proteins can also be closed through dimerization with an already closed protein, hence the signal can be rapidly propagated by a small number of activators. The reverse reaction is well-understood and proceeds through ATP-dependent remodeling of HORMA proteins from closed to open via the TRIP13 ATPase and the p31 adaptor subunit.
