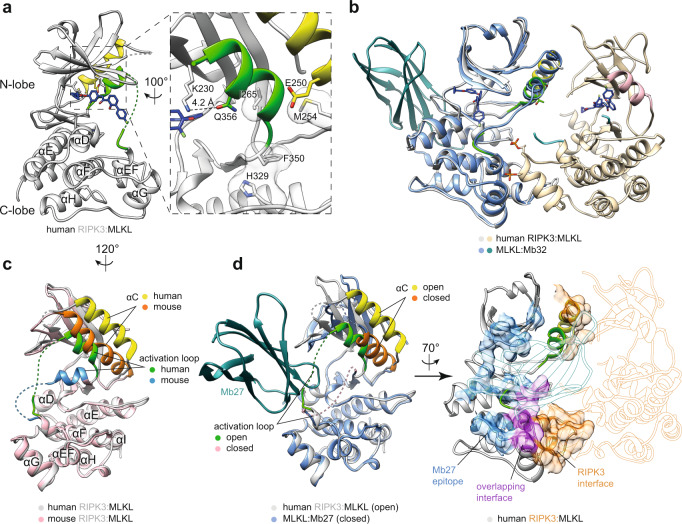
Fig. 2. Human MLKL adopts an open conformation in complex with RIPK3.
a Human MLKL pseudokinase domain (gray) from the co-crystallized complex with RIPK3. The αC helix (yellow), activation loop (green), and Compound 10 (blue sticks) are highlighted. Zoomed view on the right illustrates the open, inactive conformation adopted by MLKL. R-spine residues (gray sticks with transparent surfaces) are misaligned; the K230–E250 salt bridge interaction is disrupted by the activation loop helix supplanting the αC helix, and contributing Q356 to the interaction with K230. b Superimposition of the human MLKL (blue):Mb32 (teal) complex (PDB: 7JXU)47 with the MLKL (gray):RIPK3 (wheat) complex reported herein (PDB: 7MON). c Superimposition of the human (gray) and mouse (pink) MLKL structures from their complexes with RIPK3 from superimposing the respective MLKL protein in each complex model. Human complex (this study; PDB: 7MON); mouse, (PDB: 4M69)46. The αC helix (human, yellow; mouse, orange) and activation loop (human, green; mouse, blue) are highlighted. Viewing angle is rotated by 120° clockwise around the y-axis from the position in panel a. d A monobody (Mb27) that binds human MLKL post-release from the necrosome (left, teal; PDB: 7JW7)47 binds MLKL via a site that overlaps the human RIPK3-binding site (right; MLKL, gray; RIPK3, wheat). The MLKL αC helix (yellow) and activation loop (green) are highlighted. Left, superimposition of the human MLKL (blue):Mb27 (teal) complex with the MLKL:RIPK3 complex (same viewing angle as panel c). Right, the binding epitopes on human MLKL are shown as surfaces in the cartoon (same viewing angle as panel b); the Mb27 binding epitope is shown in blue, the RIPK3 epitope in orange and the overlapping interface in magenta. Mb27 and RIPK3 models are shown as silhouettes. All superimpositions were performed using UCSF Chimera Matchmaker.