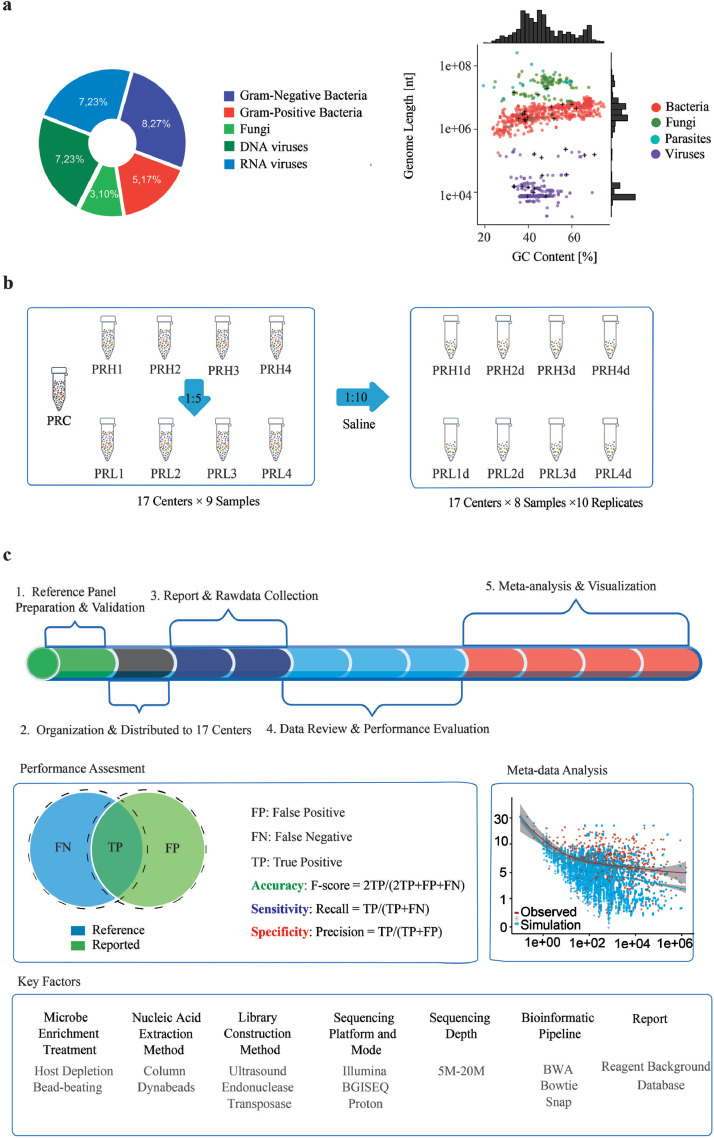
Fig. 1.
Design and overview of the study. (a) Thirty microorganisms from 19 genera were chosen to represent the diversity of microbial types (left panel), genome sizes, and GC contents (right panel). (b) Pathogen reference reagents (PRHs and PRLs) were prepared by contriving microbes at high (PRH) and low (PRL) titers with HeLa cells, respectively. PRH and PRL samples were diluted 1:10 and tested in ten replicates except for the pathogen-free PRC sample. PRC details are in the Results section. (c) Overview of study design. Numbers (1-5) order the steps of analysis. F-score, Recall and Precision metrics were used to assess the performance of accuracy, sensitivity, and specificity. Key technical factors of the workflow were explored for their impact on assay performance. See also Supplemental Tables S1–S3, and S5.