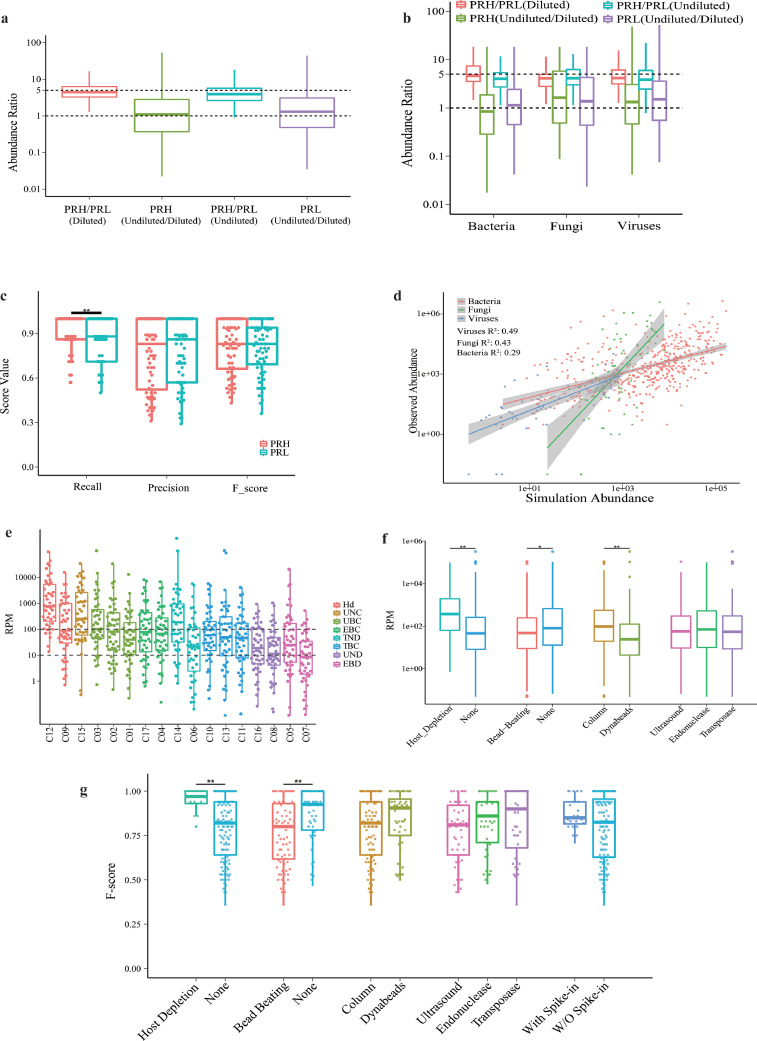
Fig. 3.
Microbial abundance and pathogen detection by metagenomics. (a,b) Observed abundance ratios between PRH and their PRL counterparts, and undiluted samples and their diluted counterparts, analyzed together (a) or by microbial type (b). (c) Correlations between relative abundance and performance (n = 136). (d) Correlations between observed and expected abundances in three microbial types, using the reads per million (RPM) of human papilloma virus within each sample as an internal control for normalization. (e,f) RPM of microbial detection varied across sites grouped by workflow, Hd: Host-deplete, UNC: Ultrasound, None, Column; UBC: Ultrasound, Bead−beating, Column; EBC: Endonuclease, Bead−beating, Column; TND: Transposase, None, Dynabeads; TBC: Transposase, Bead−beating, Column; UND: Ultrasound, None, Dynabeads; EBD: Endonuclease, Bead−beating, Dynabeads. (e) or key technical variables (f). (n = 136), *, P < 0.05, **, P < 0.01 by Wilcoxon rank sum test. See also Supplemental Figs. S3–S5. (g) Correlations between F-score and major workflow-dependent technical variables, (n = 270). **, P < 0.01 by Wilcoxon rank sum test.