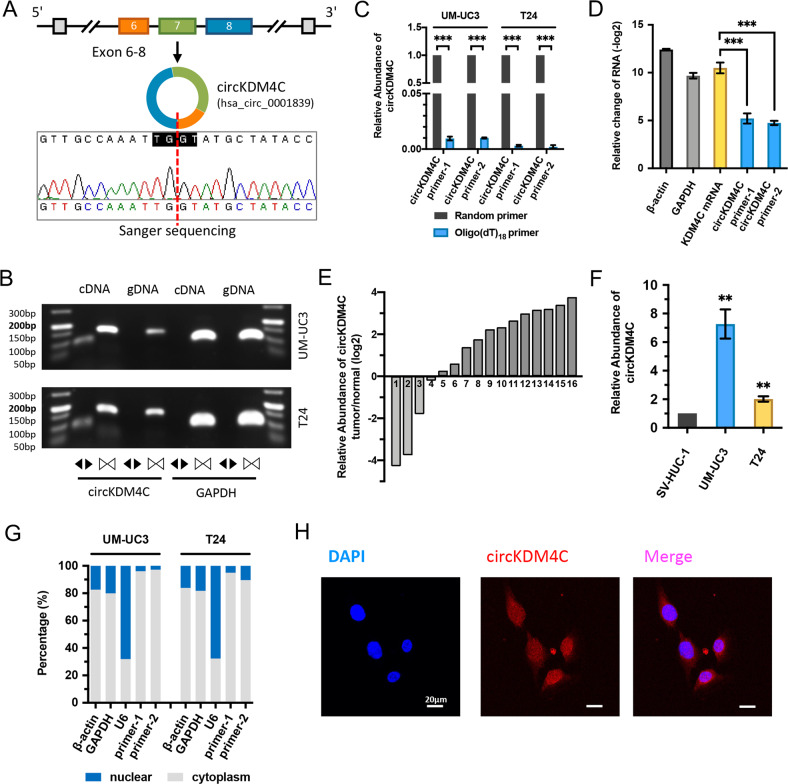
Fig. 1. Identification of circKDM4C and its expression in BCa tissues and cell lines.
A Schematic illustration shows the circularization of KDM4C exon 6–8 formed circKDM4C. Sanger sequencing was used to identify the circKDM4C back-splice junction. Red dashed line indicates the back-splicing site. B The existence of circKDM4C was confirmed in UM-UC3 and T24 cells by PCR and agarose-gel electrophoresis. Divergent primers could amplify circKDM4C in cDNA, but not in gDNA. GAPDH (105 bp) was the linear control. circKDM4C, 93 bp; KDM4C mRNA, 158 bp. C qRT-PCR analysis of circKDM4C in the cDNA constructed by random primers or oligo-dT primers. circKDM4C level was evaluated by two pairs of divergent primers. D circKDM4C and KDM4C mRNA levels after treatment with or without RNase R in UM-UC3 cells. E The expression of circKDM4C in 16 pairs of BCa tissues and adjacent normal tissues was determined by qRT-PCR. F circKDM4C levels in normal uroepithelium cell line (SV-HUC-1) and cancer cell lines (UMUC-3 and T24). G circKDM4C levels in nuclear and cytoplasmic fractions of UM-UC3 and T24 cells. H RNA FISH assay showed that circKDM4C was mainly located in the cytoplasm in T24 cells (circKDM4C probe was Cy3-labeled, the nuclei were DAPI-stained). Scale bar = 20 μm. Data are expressed as the means ± SD for n = 3. **P < 0.01; ***P < 0.001.