Fig. 3. Identification of single modified nucleosides by LC/MS/MS.
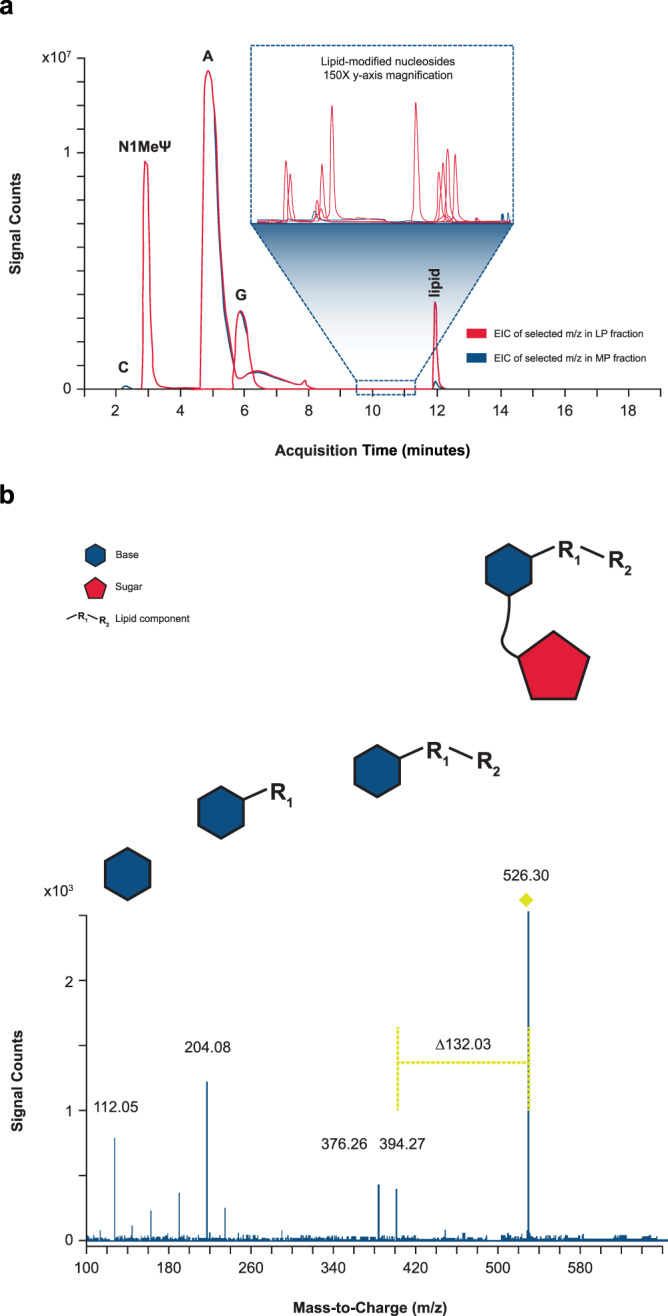
a Isolated mRNA main peak (MP) and late eluting-peak (LP) were enzymatically digested to single nucleosides and analyzed by liquid chromatography with tandem mass spectrometry (LC/MS/MS). Extracted ion chromatograms (EICs) of selected mass-to-charge ratios (m/z) corresponding to various unmodified nucleosides, lipid-modified nucleosides, and carry-over lipid in LP (red) and MP mRNA (blue) fractions are overlaid. The selected m/z for EICs include unmodified nucleosides (cytidine [C; 2.3 min], N1-methylpseudouridine [N1-MelJ.I; 3 min], adenosine [A; 5 min], guanosine [G; 6 min]), lipid (11.9 min), and several lipid-modified nucleosides (9.5–11.5 min) observed in LP mRNA fractions. 150x y-axis zoom of various low-abundant lipid-modified nucleosides eluting between 9.5 and 11 min is shown in the inset. b The precursor ion at m/z 526.30 was isolated and subjected to collision-induced dissociation. Based on fragmentation pattern, the original nucleoside was determined to be cytidine, covalently modified by one chain of the ionizable lipid prepared in the binary. The fragment ion of m/z 112.05 corresponds to the exact mass of protonated cytosine (nucleobase). The characteristic neutral mass loss of 132.03 Da, corresponds to the monoisotopic residue mass of ribose26. The fragment ion of m/z 394.27 corresponds to the lipid-modified cytosine, which further undergoes fragmentation to m/z 376.26 (loss of water) and 204.08 (at the internal ester), confirming the identity of the lipid chain. This cytidine modification is provided as a representative example, but similar characteristic neutral mass losses of 132.03 were observed for lipid modifications across all 4 nucleobases. Representative data from 4 repeat experiments are shown.
