Summary
Inflammatory bowel diseases (IBDs) are genetically complex and exhibit significant inter-patient heterogeneity in disease presentation and therapeutic response. Here, we show that mouse models of IBD exhibit variable responses to inhibition of MK2, a pro-inflammatory serine/threonine kinase, and that MK2 inhibition suppresses inflammation by targeting inflammatory monocytes and neutrophils in murine models. Using a computational approach (TransComp-R) that allows for cross-species comparison of transcriptomic features, we identified an IBD patient subgroup that is predicted to respond to MK2 inhibition, and an independent preclinical model of chronic intestinal inflammation predicted to be non-responsive, which we validated experimentally. Thus, cross-species mouse-human translation approaches can help to identify patient subpopulations in which to deploy new therapies.
Subject areas: Immunology, Computational bioinformatics, Systems biology, Transcriptomics
Graphical abstract

Highlights
-
•
MK2 kinase inhibition shows variable efficacy in different IBD mouse models
-
•
TCT and TNFΔARE mice express distinct inflammatory and MK2-responsive genes
-
•
“Response to MK2i” signature is enriched in monocytes and neutrophils
-
•
Cross-species modeling identifies patient groups potentially responsive to MK2i
Immunology; Computational bioinformatics; Systems biology; Transcriptomics
Introduction
Inflammatory Bowel Disease (IBD) – comprised of ulcerative colitis (UC) and Crohn's disease (CD) – is a chronic condition characterized by dysfunction of the mucosal barrier and uncontrolled inflammation in the gastrointestinal tract. This disease is always progressive and there is no cure. IBD can be driven by diverse genetic and environmental risk factors and causes dysregulation in multiple cell types – microbiota, intestinal epithelium, and immune system – representing a major challenge for clinicians and basic investigators trying to model and understand its etiology. Understanding the mechanisms of pathogenesis is key for the development of new treatments that attack the underlying molecular etiology of IBD. A good example of this therapeutic concept is TNF-α blocking antibodies, used in the clinic for the last two decades, and the recent approval of biologic agents that target leukocyte recruitment to the intestine or those targeting the pro-inflammatory cytokines IL-12 and IL-23 (Feagan et al., 2013; Nielsen and Ainsworth, 2013; Sandborn et al., 2009; Sands et al., 2019a, 2019b; Singh and Pardi, 2014). Unfortunately, 30–40% of patients do not respond to biologic therapy (primary non-response) and about one-third of those that initially respond become unresponsive over time (secondary loss of response) (Papamichael et al., 2015). There is an unmet need for alternative targets to treat non-responding patients and, moreover, to define which individuals will benefit from these novel therapies (Denson et al., 2019).
Preclinical studies on mouse models are key in any drug development pipeline. They provide invaluable information on mechanisms of disease, drug kinetics, specificity, and efficacy. However, one single mouse model cannot fully recapitulate all features of human disease, and this is of particular relevance in IBD, where etiology is multifactorial (contributed by both genetic and environmental factors) and patient-to-patient heterogeneity is significant. In recent years, the discipline of Translational Systems Biology has begun to systematically address the challenge of translating biological observations from preclinical disease models to patients through the development and application of computational modeling approaches (Brubaker and Lauffenburger, 2020). Our own efforts to address the challenge of cross-species translation have employed diverse methodologies including network biology, machine learning, and data-driven modeling approaches (Brubaker et al., 2019a, 2019b, 2020). We recently developed an approach for translating therapeutic responses between animal models and humans called Translatable Components Regression (TransComp-R) (Brubaker et al., 2020). TransComp-R builds a quantitative mapping between mouse and human –omics measurements that allows for inference of mouse biology predictive of human therapeutic outcomes in cases where either the mouse or human samples were not treated with the therapeutic. It is this feature of TransComp-R – quantitative modeling of cross-species differences in cases of molecular or phenotypic discrepancies – that makes it a useful tool for identifying MK2 inhibitor responses and biomarkers in IBD mouse models predictive of disease phenotypes and potential therapeutic response in treatment naive IBD patients.
In the search for novel therapeutic targets, the p38/MK2 signaling pathway has emerged as a promising candidate for the treatment of chronic inflammatory conditions, including IBD, rheumatoid arthritis, psoriasis, asthma, and chronic obstructive pulmonary disease, because it regulates cytokine production and cell migration in inflammatory conditions (Duraisamy et al., 2008; Kotlyarov et al., 1999). In response to inflammatory stimuli, MAPK kinase-3 and 6 (MKK3 and MKK6) phosphorylate and activate p38α (Enslen et al., 1998) which, in turn, phosphorylates and activates MAPK-activated protein kinase 2 (MAPKAPK2, or MK2) with which it forms a stable heterodimeric complex in the nucleus. MK2 phosphorylation exposes its nuclear export signal, resulting in export of both active p38α and active MK2 out of the nucleus (Engel et al., 1998; Enslen et al., 1998; Ter Haar et al., 2007). In the cytoplasm, MK2 phosphorylates Tristetraprolin (TTP), an RNA-binding protein that normally binds to AU-rich elements in cytokine-encoding mRNAs, targeting them for degradation in the absence of inflammatory stimuli. Phosphorylation of TTP by MK2 mediates the formation of a TTP:14-3-3 protein complex leading to TTP release from AU-rich elements (AREs) and increased cytokine mRNA translation (Cao et al., 2007; Johnson et al., 2002). Pro-inflammatory cytokines such as IL-1β, IL-4, IL-6, IL-8, GM-CSF, IFN-γ, TNF-α, and COX2 are post-transcriptionally up-regulated by MK2 by this mechanism (Duraisamy et al., 2008).
Although p38 inhibitors have been developed, these drugs were withdrawn from clinical trials due to unacceptable sideeffects and safety concerns (Dambach, 2005). As a consequence, efforts have been reoriented toward more selective therapies to block MK2-dependent signaling (Duraisamy et al., 2008). An allosteric inhibitor of the p38-MK2 heterodimer (ATI-450) has been recently developed which specifically inhibits p38-MK2 dependent signaling while sparing MK2-independent functions of p38. ATI-450 was designed to selectively bind to and inhibit the p38α–MK2 complex and it is 700x more potent in inhibiting p38α–MK2 than other p38α complexes, like p38α–PRAK or p38α–ATF2. Additionally, ATI-450 has proven to be highly selective across the human kinome, displaying >350x potency differential for inhibiting 193 kinases tested (Wang et al., 2018). ATI-450 has demonstrated efficacy in preclinical models of both rheumatoid arthritis and inflammatory bowel disease (Strasser et al., 2019; Wang et al., 2018). The question remains how to effectively deploy MK2 inhibitors in a clinical setting.
Here we aim to identify the mechanisms of response to MK2 inhibition by side-by-side comparison of MK2-dependent signaling in two mouse models of intestinal inflammation showing opposite outcomes to the MK2 inhibitor ATI-450. Further, in an attempt to translate this signaling information into relevant insights for human disease, we employed TransComp-R to investigate the potential association between the response to MK2 inhibition and relevant human disease features, particularly disease activity and response to Infliximab.
Results
Inhibition of MK2 has variable efficacy in mouse models of IBD
Given our previous demonstration of ATI-450 efficacy in an adoptive transfer model of colitis (Strasser et al., 2019), we next aimed to test ATI-450 on an alternative mouse model of IBD. The TNFΔARE mouse strain carries a deletion of the ARE in the 3′UTR Tnfa, leading to chronic over-expression of the pro-inflammatory cytokine TNF-α. TNFΔARE animals develop GI inflammation, primarily in the ileum, within 12–16 weeks after birth (Kontoyiannis et al., 1999). We chose the TNFΔARE model for several reasons. First, this mouse line develops spontaneous chronic ileitis as a result of TNF-α deregulation, recapitulating key features of Crohn's disease. Second, TNFΔARE mice are immune competent, so functional B and T cells contribute physiologically to the development of chronic ileitis. Third, our prior proteomics analysis predicted that MK2 kinase is active in this mouse model, as MK2-dependent phosphorylated targets were enriched in inflamed intestines from TNFΔARE mice (Strasser et al., 2019). Finally, previous studies have shown that MK2 genetic inactivation exacerbated inflammation in this particular mouse model (Kontoyiannis et al., 2002), therefore, we predicted that the inhibitor would not be effective in this model.
We first evaluated by western blotting the phosphorylation of MK2 and its immediate downstream target Hsp27. Although phospho-MK2 was not upregulated upon inflammation, phosphorylation of Hsp27, a direct MK2 substrate, was significantly up-regulated in inflamed tissues relative to controls, suggesting that inflammation is associated with up-regulation of MK2 activity (Figure 1A). As expected, ATI-450 stabilized the levels of total MK2 and reduced Hsp27 phosphorylation, indicating that it inhibits signaling through the p38/MK2 pathway (Figure 1A).
Figure 1.
Inhibition of MK2 has variable efficacy in mouse models of IBD
(A) Western blots of ileum tissues from TNF+/ΔARE mice and WT littermates treated with MK2 inhibitor ATI-450 or vehicle for 3 weeks. Dashed line indicates where irrelevant lanes have been digitally removed. Band quantification plots are shown on the right. Data are represented as mean ± SEM, ∗p value < 0.05; ∗∗p value < 0.01; ∗∗∗p value < 0.001; ∗∗∗∗p value < 0.0001 (two-way ANOVA test).
(B) Representative H&E images of ilea from TNF+/ΔARE mice and colons of TCT mice treated with ATI-450 or vehicle. Scale bar 200 μm.
(C) Pathology scores of TNFΔARE and TCT mouse models after ATI-450 treatment. Scoring system summarizes the extent and severity of leukocyte infiltration, epithelial changes (crypt hyperplasia and goblet cell loss) and mucosal architecture (villi blunting, crypt loss). Data are represented as mean ± SEM (multiple t-test vehicle vs. ATI-450, ∗∗∗ < 0.0001).
(D) 32-plex Luminex quantification of murine cytokines/chemokines in ilea tissues of TNF+/ΔARE mice and littermate controls after vehicle or ATI-450 treatment. Normalized z-scored values by analyte are represented. ∗p value<0.05; ∗∗∗p value<0.001; ns, non-significant (Student's t-test vehicle vs. ATI-450).
At 16 weeks of age, TNFΔARE/+ mice and WT controls were treated with ATI-450 following the same protocol previously demonstrated to be efficacious in the T cell transfer (TCT) model. Briefly, ATI-450 was formulated in the chow at 1000ppm and given to animals ad libitum for up to 3 weeks. After 3 weeks of treatment, and even after confirmed targeting of MK2 signaling pathway (Figure 1A), TNFΔARE/+ mice showed no evidence of improvement after ATI-450 treatment. Villi blunting, crypt loss, and leukocyte infiltration were not significantly improved after ATI-450 treatment and therefore pathology scores (which semi-quantitatively summarize key aspect of chronic ileitis) were not significantly different between treated and untreated mice (Figure 1B). This finding was in clear contrast to ATI-450 treatment outcome in the TCT model of colitis. In this model and consistent with our prior work, administration of ATI-450 to animals with colitis resulted in improvement of pathology scores, with significant reduction in the levels and extent of leukocyte infiltration, crypt dysplasia, and goblet cell loss (Figure 1B).
We previously reported that ATI-450 induces a significant down-regulation of proinflammatory cytokines and chemokines in the TCT model, including G-CSF and MIP2 (Strasser et al., 2019). We next explored whether cytokine levels were similarly altered after MK2 inhibition in the TNFΔARE model. Twenty-three cytokines and chemokines were surveyed on ileal lysates by Luminex. As expected, TNFΔARE/+ mice showed a clear upregulation of most of the pro-inflammatory cytokines compared to WT littermates, including G-CSF, GM-CSF, IL1-α, IL1-β, IL-6, IL-10, IL-12 (p70), IL-15, IL-17, IP-10, KC, MCP-1, MIP1-α, MIP1-β, MIP-2 and TNF-α (Figure 1C, Table S1). However, most – with the exception of IL-12p40 and IL-15 – were not affected by ATI-450 treatment (Figure 1C). Altogether, our data indicate that, although MK2 is activated during inflammation in the TNFΔARE model and ATI-450 inhibits MK2, inhibition of the pathway is not associated with an abrogation of cellular or molecular inflammatory phenotypes.
TNFΔARE and TCT models exhibit inflammation responses with unique features
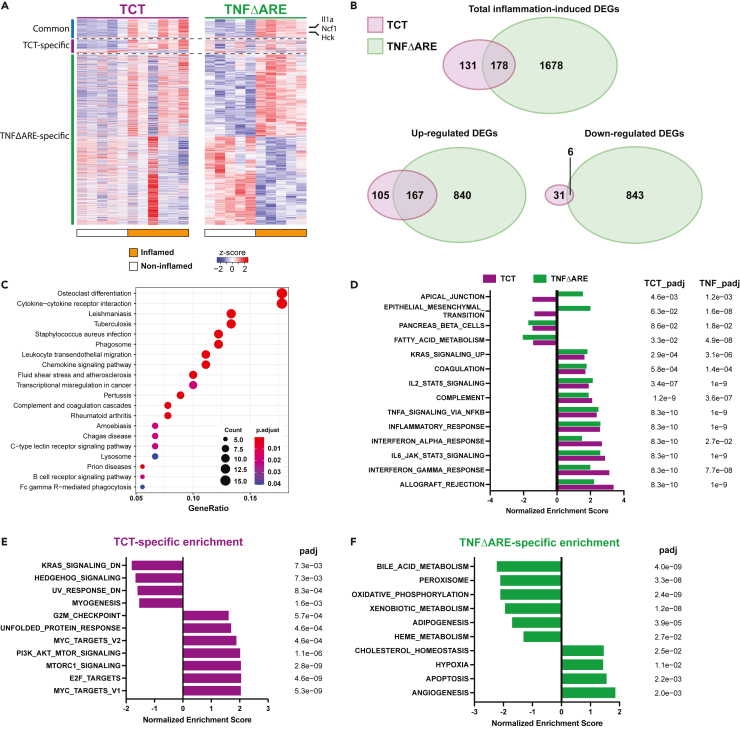
To understand differential responses to MK2 inhibitor, we performed transcriptomic profiling of inflamed tissues and controls from the TCT and TNFΔARE mouse models (Tables S2and S3). Hierarchical clustering, principal component analysis (PCA), and MA plot demonstrated clean clustering of samples based on their inflammation status, with even read distribution for both models (Figure S1). There were hundreds of differentially expressed genes (DEG, based on adjusted p value < 0.05 and log-fold change >0.25) identified in inflamed tissue from each model (Figure 2A). Interestingly, there was a clear difference in the number of DEG (309 in the TCT model and 1,856 in the TNFΔARE model), likely due to the inter-animal variability of the different models, with TNFΔARE (a genetic model of inflammation) being more consistent from animal-to-animal than TCT (inflammation induced by adoptive transfer). Comparative analysis of DEGs from both models revealed an overlap of only 178 genes, 173 of which changed in the same direction in the two models (167 up-regulated and 6 down-regulated, Figure 2B). We surmised that these 173 DEGs likely reflect inflammatory processes that are shared by the two models, as they include up-regulations of genes like Il1a, Ncf1, and Hck (Lowell and Varmus, 1994; Segal et al., 2002; Zheng et al., 2013). KEGG and GO enrichment analysis of the 173 overlapping DEGs identified many general inflammation-related pathways, including cytokine-cytokine receptor interaction, phagosome, chemokine signaling pathway (Figure 2C, Tables S4 and S5), confirming that the shared DEGs represent an inflammation signature that is shared between these two models of IBD. Additionally, 1,923 DEGs were identified upon unifying the DEGs from both models (detected in both, significant in either model, Table S6). 75% of these genes (1451/1923) were perturbed in the same direction upon inflammation in both models, further underscoring the general inflammatory program.
Figure 2.
TNFΔARE and TCT models exhibit inflammation responses with unique features
(A) Transcriptomic profiling identified 309 and 1856 DEGs during inflammation in the TCT and TNFΔARE models (adjusted p value < 0.05, LFC > 0.25, Wald test with Benjamini-Hochberg correction), with 178 overlapping.
(B) A moderate number of DEGs (178/309) from TCT model overlap with DEGs from TNFΔARE, most of which are dysregulated in the same direction (173/178).
(C) KEGG Analysis of the 173 overlapping DEGs of inflammation yielded enrichment of general inflammation-related pathways, validating TCT and TNFΔARE as models of intestinal inflammation.
(D) Hallmark gene set GSEA of TCT and TNFΔARE transcriptomic profiles excluding the 173 overlapping DEGs revealed 14 overlapping pathways, 12 of which are enriched in the same direction. Notably, several inflammation-related pathways are enriched in both models.
(E) TCT-specific GSEA pathway enrichment.
(F) TNFΔARE-specific GSEA pathway enrichment. Note that mTORC1 signaling and PI3K-AKT-mTOR signaling pathways are enriched specifically in TCT (p value < 0.05, adjusted p value < 0.1, adaptive multi-level split Monte-Carlo with Benjamini-Hochberg correction).
See also Figure S1.
Next, we focused on model-specific DEGs. Of the 309 genes differentially expressed during colitis in the TCT model, 131 (42.4%) were specific to this model. To identify model-specific features, we initially performed KEGG and GO enrichment analyses using overlapping DEGs and model-specific DEGs. Given the much larger number of TNFΔARE specific DEGs compare to those of the TCT model (1,676 vs 131, Figure 2B), more enrichment terms were identified using the TNFΔARE data (Figure S2A, Tables S7, S8, S9, and S10). This considerably limited the comparability of these enrichment analyses. To circumvent the issue, we performed GSEA on the complete TCT and TNFΔARE transcriptomic profiles, excluding the 173 overlapping DEGs (Tables S11 and S12). Enriched pathways from the Hallmark gene-set were identified (p value < 0.05, adjusted p value < 0.1). Fourteen pathways were commonly enriched between TCT and TNFΔARE, including many inflammation-related pathways such as allograft rejection and interferon responses (Figure 2D). This, again, validated that the two models share general underlying inflammatory mechanisms. Interestingly, TCT-specific pathways included enrichment of MYC target V1/V2, PI3K-AKT-mTOR signaling, and mTORC1 signaling, and the depletion of the KRAS signaling down signature (Figure 2E), suggesting activation of growth factor receptor signaling in the TCT model of inflammation. Conversely, TNFΔARE-specific pathways included predominantly metabolism-related gene sets, such as the enrichment of cholesterol homeostasis, and depletion of adipogenesis, bile acid metabolism, oxidative phosphorylation, and fatty acid metabolism (Figure 2F). These data suggest that despite sharing a general mechanism of inflammation, the two IBD models exhibited unique aspects of transcriptomic changes that potentially contributed to their differential responses to MK2 inhibitor treatment.
Transcriptomic signatures of MK2 inhibitor response
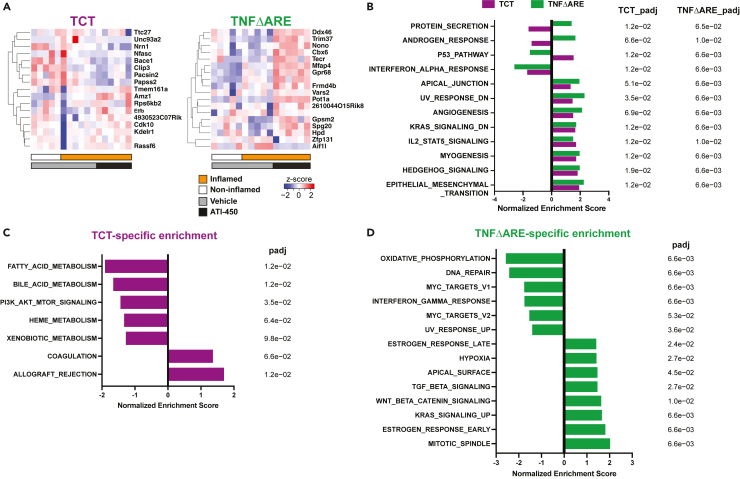
In addition to unique inflammation signatures, model-specific effects of MK2 inhibitor might also contribute to their different drug responses. To characterize the molecular responses to MK2 inhibitor treatment in both models, animals with and without inflammation were treated with ATI-450 for 16 h prior to harvesting intestinal tissue for transcriptomic profiling (Tables S13 and S14). The acute treatment timepoint was chosen to ensure detection of the immediate effects of MK2 inhibition, rather than transcriptional changes tied to the tissue inflammatory state. However, this also decreased the detected signal intensity of MK2 inhibition, contributing to the lack of clear clustering of MK2 inhibitor treated samples from the controls (Figures S1A and S1C). In TCT and TNFΔARE animals with inflammation, we identified 17 and 18 genes that were altered in expression following MK2 inhibitor treatment (Figure 3A, adjusted p value < 0.1, log-fold change >0.25). No overlap was found between MK2 targets in the two models, suggesting model-specific functions of MK2. To further illustrate the model-specific features of MK2 signaling, we performed GSEA using the Hallmark gene-set on transcriptomic profiles of inflamed samples treated with vehicle or ATI-450 (Tables S15 and S16). We identified 12 pathways that were significantly enriched and shared between the two models (Figure 3B, p value < 0.05, adjusted p value < 0.1). Of these, 9 pathways changed in the same direction, including angiogenesis and interferon-alpha response. The other 3 pathways were enriched in opposite directions, including the p53 pathway, androgen response, and the protein secretion pathway. We also identified a variety of pathways that were uniquely enriched in the TCT and TNFΔARE models following MK2 inhibitor treatment (Figures 3C and 3D). Interestingly, the PI3K/AKT/mTOR pathway which was specifically upregulated upon inflammation in the TCT model but not in TNFΔARE mice (Figure 2E), was also uniquely down-regulated by MK2 inhibitor in the TCT model (Figure 3C).
Figure 3.
MK2 inhibitor elicits model-specific transcriptomic changes
(A) DEGs were identified comparing transcriptomic profiles of inflamed TCT and TNFΔARE mice with vehicle or ATI-450 (adjusted p value < 0.1, LFC > 0.25, Wald test with Benjamini-Hochberg correction).
(B) Hallmark GSEA analysis of data from ATI-450 treated animals showed common unidirectional changes in 9 pathways between TCT and TNFΔARE animals. Three pathways with opposing enrichment between the two models were also identified (p value < 0.05, adjusted p value < 0.1, adaptive multi-level split Monte-Carlo with Benjamini-Hochberg correction). GSEA analysis also identified model-specific pathway enrichments for (C) TCT and (D) TNFΔARE. Note that PI3K-AKT-mTOR signaling is uniquely down-regulated in inflamed TCT animals treated with ATI-450.
See also Figure S1.
In order to identify the downstream effectors targeted by the MK2 inhibitor that could be mediating the resolution of inflammation, we examined the leading edge genes driving the enrichment of the Hallmark PI3K/AKT/mTOR pathway in inflamed TCT mice (Figure 2E) and MK2i-treated TCT mice (Figure 3C). We identified a total of 26 genes present in both leading edge gene sets, which we defined as the MK2 target genes, several of which were related to cytokine receptor signaling (Tnfrsf1a, Mknk1, Myd88, Dapp1, Stat2, Tbk1, Ripk1, Ptpn6), NF-κB signaling (Nfkbib, Trib3, Ikbke, Mapk8, Map2k3), membrane-associated GTPases signaling (Vav2, Raf1, Rac1, Grk2, Akt1, Ywhaz, Pdk1, Arhgdia), Wnt signaling pathway (Csnk2b, Gsk3a), actin cytoskeleton and cellular trafficking (Actr3, Ap2m1) and cell cycle regulation (Cdk1).
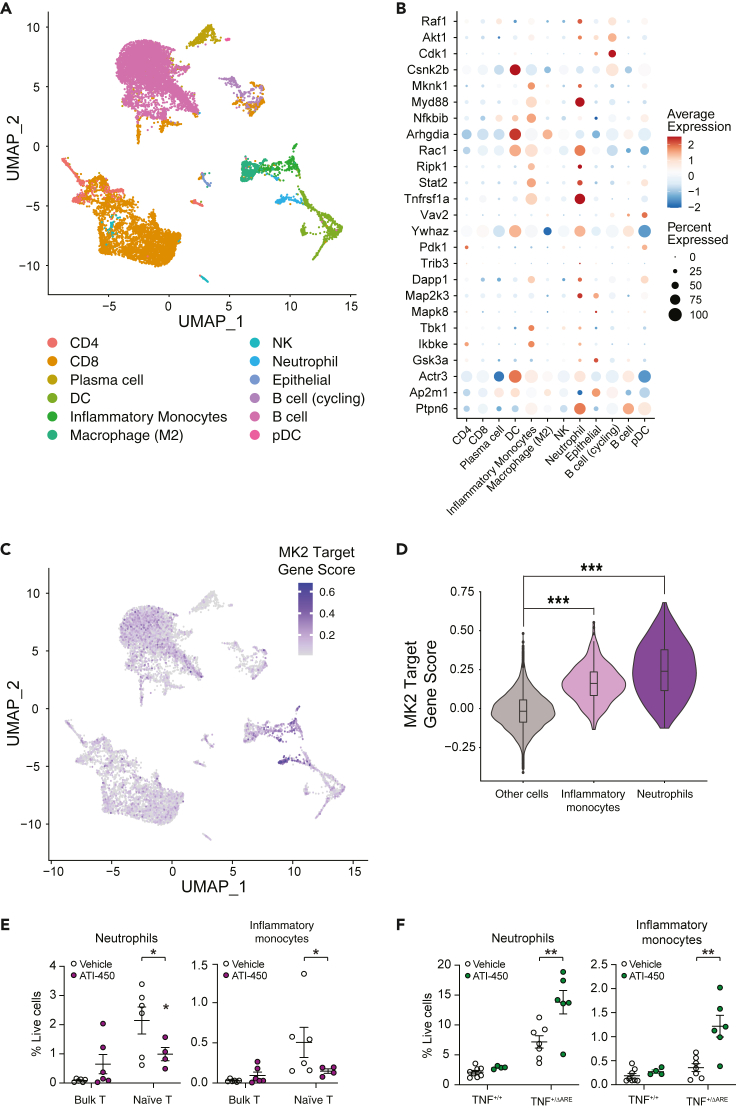
MK2i target gene expression is enriched in intestinal inflammatory monocytes and neutrophils
MK2 is expressed ubiquitously and little is known about the exact role of this kinase in driving inflammation in the intestinal mucosa, for example which cellular compartments are contributing to inflammation in an MK2-dependent manner. In order to gain more insight into the pro-inflammatory role of MK2, we utilized a published single-cell RNA-Seq (scRNA-Seq) dataset from mouse intestine to analyze the cellular localization of the MK2 target genes. The dataset consisted of CD45-enriched MHCIIfl/fl (WT) mouse intestinal cells at homeostasis and 4 days post Helicobacter polygyrus infection (Biton et al., 2018). Twelve distinct clusters of cells were identified, primarily composed of immune cells (Figure 4A). Cell composition significantly changed at 4 days post-infection, with a decrease of B cells and increase of CD8+ T cells, neutrophils, and inflammatory monocytes (Figures S3A and S3B). Gene set variation analysis (GSVA) revealed enrichment of inflammation-related pathways in inflammatory monocytes and neutrophils, confirming the inflammatory status of the intestine 4 days post infection (Figure S3C). All but one of the MK2 target genes were detected, most of which were commonly detected and highly expressed in inflammatory monocytes (Figure 4B). MK2 target gene score was calculated for each cell to identify potential candidate cell populations involved in MK2 inhibition response. Inflammatory monocytes and neutrophils scored significantly higher than other cell types (Figures 4C and 4D), implicating the potentially significant role for MK2 activation these two cell types in driving inflammation. Further validation using a separate scRNA-Seq dataset from the inflamed ileum of Crohn's Disease patients confirmed the enrichment of MK2 target gene signature in the inflammatory monocyte and neutrophil compartment (Figure S4) (Martin et al., 2019).
Figure 4.
MK2 target gene expression is enriched in murine inflammatory monocytes and neutrophils
(A) Single cell UMAP clustering of CD45+ enriched cell populations from the small intestine of mice at homeostasis or 4 days post-infection of H. polygyrus. Raw data from (Biton et al., 2018) (GEO: GSE106510). Original cell annotations of the 12 cell types were used.
(B) MK2 target genes are commonly detected and highly expressed in inflammatory monocytes and neutrophils.
(C) UMAP display of MK2 Target gene score for all cells demonstrates high MK2 target gene signature in inflammatory monocytes and neutrophils.
(D) Overlay of violin plot and box plot shows that MK2 target gene expression is significantly enriched in intestinal inflammatory monocytes and neutrophils (∗∗∗ p value < 2.2 × 10−16, Wilcoxon rank-sum test).
(E) FACS-mediated quantification of myeloid cell recruitment to colonic (TCT) or (F) ileal (TNFΔARE) mucosa in mice after ATI-450 or vehicle treatment. Myeloid cells are defined as CD45 + CD11b+ cell. Neutrophils and Inflammatory Monocytes are defined within myeloid cells gate based on GR1 and Ly6C surface staining. Data are represented as mean ± SEM. ∗ p value < 0.05; ∗∗ p value < 0.01 (Student's t-test vehicle vs. ATI-450).
See also Figures S3 andS4.
In order to better understand the how inhibition of MK2 affects neutrophils and monocytes in the context of colitis, we characterized the immune cell populations recruited to the intestinal mucosa in the TCT model, both during inflammation and after MK2 inhibition. ATI-450 significantly reduced the number of neutrophils and inflammatory monocytes recruited to intestinal mucosa in the TCT model (Figure 4E). We also measured myeloid recruitment after MK2 inhibition on the non-responder model TNFΔARE mice. In clear opposition to TCT mice, MK2 inhibition resulted in a significant increase of both neutrophil and inflammatory monocytes recruitment to intestinal mucosa of TNFΔARE mice (Figure 4F). In sum, our transcriptomic analysis indicates that MK2 inhibitor likely targets the myeloid compartment (neutrophils and inflammatory monocytes) to resolve inflammation in the TCT mouse model.
Integrating and translating mouse model signatures into relevant clinical information
We were able to identify a potential mechanism of response to MK2 inhibition by transcriptomic analysis in the murine models; however, identification of effective therapies for IBD patients requires translating mouse models findings into relevant human clinical information. In translating the MK2 inhibition observations from mouse models to human IBD, we sought to consider two clinical phenotypes in humans. First, we asked whether a gene signature that distinguished MK2 responsive and non-responsive mice could be used to predict human disease status as an initial test of translational potential. We then asked whether the genes distinguishing MK2 responsive and non-responsive mice could be used to position MK2 inhibition therapy to patients with clinical need, specifically those patients resistant to the anti-TNF therapy, Infliximab. Although there are other differences between TCT and TNFΔARE mice likely not related to MK2i response, we sought to assess whether the observable gene differences were translationally consequential for humans. Despite the apparent differences between the systems, MK2i response in mice and anti-TNF resistance in humans, bridging such systems is an important goal of preclinical-to-clinical translation.
We hypothesized that combining information from multiple mouse models might recapitulate interpatient heterogeneity and therefore provide a platform to apply computational modeling approaches to translate our findings to human IBD populations, allowing us to identify patient subgroups that resemble one mouse model versus another. Here, we sought to identify transcriptional signatures that could distinguish TCT mice (“MK2i responsive”) from TNFΔARE mice (“MK2i non-responsive”) and predict relevant human IBD features, such as disease state and response to anti-TNF-α therapy (i.e. Infliximab). We used published human IBD gene expression datasets that include data from ulcerative colitis (UC, n = 24), colonic Crohn's disease (cCD, n = 19), and ileal Crohn's disease (iCD, n = 18) patients with colonic (n = 6) and ileal (n = 6) controls (Arijs et al., 2009).
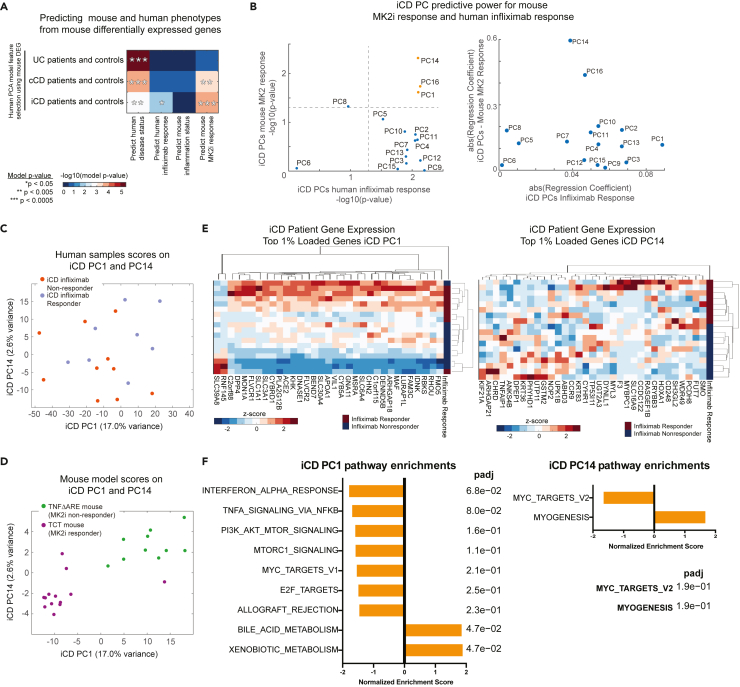
By comparing TCT (pooling controls and inflamed) vs TNFΔARE (controls and inflamed), we found 3,268 genes differentially expressed between the two models of intestinal inflammation (FDR q < 0.05 Table S17). This gene set was used to select genes in the human gene expression datasets from which we constructed three principal component analysis (PCA) models for subsets of UC patients, cCD patients, and iCD patients with controls. For each human IBD subtype PCA model, we performed two principal component regressions (PC-R), regressing human scores on human PC's against either disease state or response to Infliximab. These human PC-R models were used to identify those human PC's, comprised of genes differentially expressed between the TCT and TNFΔARE mouse models that best predicted human disease and therapeutic response phenotypes. However, these patients were never treated with an MK2 inhibitor and MK2i response was unknown for these patients.
In order to identify human IBD gene signatures predictive of MK2i response, we employed a cross-species modeling approached previously developed by our group called Translatable Components Regression (Brubaker et al., 2020). For each IBD-subtype PCA model, we projected the TCT and TNFΔARE mouse RNA-seq profiles onto the human PC's, causing relative differences between mouse models were expressed in terms of positions in human PC space. We then trained two regression models using the mouse samples and their projections in human PC space, first predicting mouse inflammation status (Y: inflamed vs. uninflamed) and then predicting mouse model type (Y: TCT or TNFΔARE) to identify human PC’s predictive of mouse phenotypes (Tables S18, S19, and S20, Figure 5A). By combining the results of the PC-R (human PC's predictive of human phenotypes) and TransComp-R (human PC's predictive of mouse phenotypes), we were able to identify PCs and gene signatures predictive of both mouse and human IBD phenotypes. For colonic CD, we identified human PC's predictive of at least one human and mouse phenotype. Though we identified human PC's predictive of UC disease status, these PC's were not predictive of either mouse inflammation status or mouse model type. The most predictive PCs of patient disease activity in cCD did not coincide with the predictive PCs of mouse inflammation, therefore, these cCD PCs may not be strongly translatable between mice and humans (Figure S5A). By contrast, the PC-R and TransComp-R models identified iCD PCs that were predictive of disease status and Infliximab response in iCD patients, as well as the type of mouse model. The most predictive PCs were PC1, PC14, and PC16 (Figures 5B and S5B), where the axis of separation between TNFΔARE and TCT mice resembles the axis of separation between Infliximab responsive and resistant patients (Figures 5C and 5D). The comparisons of cross-species predictive power are inferred by comparing the significance of the linear model regression coefficients for the iCD PCs when used to predict human phenotypes in a PC-R model or mouse phenotypes in a TransComp-R model. These results indicate that the genes specifying iCD PC1, PC14, and PC16 were the best candidates for therapeutic response signatures translatable from mouse models to humans and the genes driving separation along PC1 and PC14 separate patients Infliximab response (Figure 5E, Table S21). These three human PCs together account for 22.1% of the total variance in the iCD transcriptomics data, with PC1 the major contributor (17% of the human transcriptomics variance explained).
Figure 5.
Integrated translational modeling of murine IBD transcriptomics
(A) Regression model (TransComp-R and PC-R) significance for identifying human IBD subtype principal components predictive of human disease status (PC-R), human response to Infliximab (PC-R), or mouse MK2 inhibitor (MK2i) treatment response or inflammation status (TransComp-R) using genes differentially expressed between TCT and TNFΔARE mouse models.
(B) Predictive power of human iCD PCs for human Infliximab response and mouse MK2i treatment response as denoted by regression coefficient p values and regression coefficient magnitudes.
(C) Human samples plotted in the iCD PC space along iCD PC1 and PC14.
(D) Mice plotted in the iCD PC space along iCD PC1 and PC14.
(E) Gene expression data from iCD patients for genes with top 1% loading coefficient magnitudes on iCD PC1 and PC14.
(F) Pathway enrichment analysis of iCD PC1, PC14, and PC16. Significant pathways of iCD PC1 and iCD PC14 are shown. No pathways were significantly loaded along iCD PC16.
See also Figure S5.
In order to better understand the biological relevance of these computational models, we performed GSEA analysis of the gene loadings of the predictive PC's and identified significantly enriched pathways on PC1 and PC14 predictive of human Infliximab response and the inflammatory context/MK2i responsiveness of the two mouse models. The GSEA hallmark pathways bile acid metabolism, xenobiotic metabolism, MYC, and PI3K/AKT/mTOR signaling were significantly enriched in PC1 gene loadings, along with MYC signaling and myogenesis on PC14, suggesting that these pathways may be translatable biomarkers of Infliximab resistance (Figure 5F). These results indicate that the same gene signatures predictive of iCD response to anti-TNF-α therapy are also predictive of the type of IBD mouse model (TCT vs. TNFΔARE), a variable equivalent in the model to MK2i responsiveness.
The gene signature differentiating mouse models can predict the response to MK2 inhibitor
We have used the transcriptomic differences between two independent mouse models to train computational models that predict whether an IBD patient resembles the TNFΔARE or TCT mouse model, which differ in their responsiveness to inhibition of MK2. In order to validate the predictiveness our computational models, we next sought to investigate how well the models could infer the response to ATI-450 on a third mouse model of intestinal inflammation, SAMP1/YitFc. The SAMP1/YitFc mouse model is a substrain of AKR/J that was generated by selective breeding for a senescence-associated phenotype (Matsumoto et al., 1998). Like the TNFΔARE model, SAMP1/YitFc mice develop chronic ileitis without chemical or immunological manipulation (Pizarro et al., 2011). In contrast to the TNFΔARE model, the genetic basis for inflammation in the SAMP1/YitFc model is unknown. When reaching 20 weeks of age, SAMP1/YitFc mice exhibit clear signs of ileitis, including transmural leukocyte infiltration, villi blunting, crypt loss, and muscle layer hyperplasia, which is accompanied by stricturing disease.
We measured MK2 pathway activation in the SAMP1/YitFc mice. Both phospho-MK2 and phospho-Hsp27 are present in ileal tissue samples, indicating that the pathway is active. However, there is not a significant upregulation of phospho-MK2 or phospho-Hsp27 in ileal samples of inflamed SAMP1/YitFc mice compared to AKR controls, which indicates that the pathway is active in the ileum tissues independently of the inflammatory state. We further confirmed that ATI-450 was able to downregulate MK2 signaling in this mouse model, illustrated by the stabilized MK2 protein levels and downregulated phosphorylation of Hsp27 in both SAMP1/YitFc mice and AKR controls (Figure 6A).
Figure 6.
The gene signature differentiating mouse models can predict the response to MK2 inhibitor
(A) Western blots of ileum tissues from SAMP1/YitFc mice and AKR controls treated with MK2 inhibitor ATI-450 or vehicle for 2 weeks ad libitum.
(B) Projection of transcriptomics data from SAMP1/YitFc mouse samples onto human iCD PC space, along with the MK2 inhibitor responsive (TCT) and non-responsive (TNFΔARE) mouse samples.
(C) Hierarchical clustering of loading coefficients on PC1, PC8, PC14 and PC16 of SAMP1/YitFc, TCT, and TNFΔARE mice. Note that SAMP1/YitFc mice cluster with TNFΔARE mice.
(D) Representative H&E images of ilea from SAMP1/YitFc mice and AKR controls treated orally with either ATI-450 or vehicle. Scale bar, 200μm.
(E) Pathology scores of inflamed SAMP1/YitFc mice and AKR controls after ATI-450 treatment. Scoring system summarizes the extent and severity of leukocyte infiltration, epithelial changes (crypt hyperplasia and goblet cell loss), mucosal architecture (villi blunting, crypt loss) and muscle layer hypertrophy. Data are represented as mean ± SEM. None of the comparisons were statistically significant (Student's t-test vehicle vs. ATI-450).
Next, we obtained publicly available gene expression data from epithelial stem cells in the small intestines of SAMP1/YitFc mice (Buttó et al., 2020) and sought to predict the MK2i responsiveness of this mouse model using the expression data from the genes used to train our predictive models. Although the cells from the SAMP1/YitFc mice were not whole-tissue like the other mice, this dataset was the most similar intestinal gene expression dataset to our mice, making it the best-available dataset for the SAMP1/YitFc mice to test our model. We projected the SAMP1/YitFc expression data onto iCD PC's associated with MK2i response in mice to preserve the translational relevance of this prediction, these being iCD PC1, PC8, PC14, and PC16 (Figure 6B). Hierarchical clustering of the mouse projection scores on the iCD PCs associated with MK2i response indicated that SAMP1/YitFc mice projected most similarly to TNFΔARE mice on human PCs, suggesting that these mice, like TNFΔARE, would be resistant to MK2 inhibition with ATI-450 (Figure 6C).
Because the cell types profiled for the SAMP1/YitFc mice differed from those of the TNFΔARE, we sought to test whether the resemblance of gene expression from SAMP1/YitFc intestinal crypts was sufficient to predict MK2i response. We therefore dosed inflamed SAMP1/YitFc animals and age-matched AKR controls with ATI-450 formulated in the chow for 2 weeks ad libitum. At the end of the treatment, mice were euthanized, and ileum samples processed for histologic evaluation (Figure 6D). Blinded scoring of ileum sections demonstrated that MK2 inhibition did not affect any of the histologic effects of ileitis – villi blunting, crypt loss, or thickening of the muscle layer – which was translated into no significant improvement of pathology scores (Figure 6E). The lack of efficacy of ATI-450 in this mouse model validated our computational prediction, therefore indicating that our PC models effectively predict whether a given mouse model is responsive to MK2 kinase inhibition.
Discussion
MK2 is a master regulator of pro-inflammatory cytokines, such as TNF-α, IL-1β and IL-6, which are critical mediators of chronic inflammatory diseases. Not surprisingly, MK2 targeting has been postulated as a potential therapeutic strategy for autoimmune and inflammatory diseases like arthritis, atherosclerosis, Alzheimer disease and IBD (Duraisamy et al., 2008; Fiore et al., 2016). In fact, MK2 inhibition alleviates inflammation in several preclinical models of intestinal and joint inflammation (Strasser et al., 2019; Wang et al., 2018). However, not all models of IBD respond to MK2 inhibition. The efficacy of MK2 inhibition in the context of intestinal inflammation has remained controversial. While MK2 inhibition has shown pre-clinical efficacy in the T cell transfer model and MK2 KO mice exhibit attenuated DSS-induced colitis, genetic inactivation of MK2 exacerbates inflammation in the TNFΔARE spontaneous of ileitis model (Li et al., 2013; Strasser et al., 2019; Zhang et al., 2020). When crossed into whole-body MK2 KO mice, the double mutant mice showed early formation of multiple granulomas and extensive lymphocytic aggregates in the lamina propria that was also associated with high incidences of mortality from the age of 8 weeks onwards (Kontoyiannis et al., 2002). Using a clinical MK2 inhibitor (ATI-450), our studies corroborate that disruption of MK2 signaling is not therapeutically effective in the TNFΔARE model (Figure 1).
In the search for the mechanism through which MK2 inhibition can resolve inflammation in some contexts, we performed a side-by-side comparison of the transcriptomes of two models of intestinal inflammation with opposite outcomes to inhibition: TCT (responder) and the aforementioned TNFΔARE (non-responder). Both models recapitulate key features of human Crohn's disease: chronic intestinal inflammation with transmural infiltration of leukocytes and predominant Th1 responses, even though the site of manifestation is predominantly colonic in the TCT and ileal in TNFΔARE (Pizarro et al., 2003). We defined a set of genes – enriched for PI3K/AKT/mTOR pathway target genes – that are associated with the positive response to MK2 inhibition (Figures 2E and 3C). This observation is consistent with our prior demonstration that mTOR signaling is activated in the colons of TCT mice and that inhibition of the pathway leads to resolution of inflammation. During inflammation, mTOR is activated in intestinal epithelial cells, where it promotes an undifferentiated cellular state that enhances inflammatory cytokine secretion (Lyons et al., 2018). Our analysis of single cell RNAseq data located MK2 target genes, which contribute to the leading edge of PI3K/AKT/mTOR hallmark pathway, to myeloid cells, particularly neutrophils and pro-inflammatory monocytes (Figure 4). Multiple MK2 roles in myeloid cells have been recently described: MK2 promotes reactive oxygen species production by neutrophils, regulates monocyte recruitment, drives TNF-α and IL-1β expression on pro-inflammatory macrophages, and regulates alternative polarization of M2 macrophages (Gaestel et al., 2009; Limbourg et al., 2015; Suarez-Lopez et al., 2018; Sun et al., 2018; Zhang et al., 2020). Our results suggest that MK2 inhibition is alleviating inflammation in the TCT model because it is able to regulate inflammatory monocytes and neutrophil function. Indeed, ATI-450 treatment results in a significant decrease of both inflammatory monocytes and neutrophils to colonic mucosa of TCT mice, but not in the ileal mucosa of TNFΔARE animals (Figures 4E and 4F). These results are largely consistent with previous cross-species modeling efforts in UC that highlighted the importance of neutrophil functions and inflammatory pathways in biologic therapy resistance (Czarnewski et al., 2019).
IBD is highly heterogeneous and one single preclinical model cannot recapitulate all of the disease features that are seen in patients. Thus, it is perhaps not surprising that mouse models with distinct disease etiologies would exhibit dissimilar responses to a given perturbation, just as therapeutic responses vary in genetically diverse human patients. In an attempt to translate our mouse findings into relevant information for IBD, we made use of TransComp-R, which is based on the hypothesis that combining biological variation from multiple animal models better recapitulates human disease variation than a single animal model. Based on this hypothesis, we built computational models to predict human clinical features using genes differentially expressed between the TCT and TNFΔARE mouse models. The gene signature that predicted clinical features in ileal Crohn's disease coincided with the gene signature associated with a positive response to MK2 inhibitor (TCT signature) and, furthermore, this signature predicted the efficacy of the inhibitor on an independent mouse model of intestinal inflammation. Interestingly, the gene signature of sensitivity to MK2 inhibitor coincides with the one predictive of ileal CD patient's non-responsiveness to anti-TNF-α therapy. This finding has the potential to be translated between IBD mouse models and humans since this signature was implicated on iCD PC1, a PC explaining a large portion of patient-to-patient variation. Although further studies are needed to fully assess this finding in humans, our results indicate that patients that are resistant to anti-TNF-α may be responsive to MK2 inhibition therapy.
Systems approaches to cross-species modeling have taken many forms in recent years in an effort to address the important challenges of preclinical-to-clinical translation. Our previous work has employed a variety of supervised and semi-supervised modeling approaches to analyze similarly sized mouse and human cohorts and to the present work to make and validate cross-species predictions of disease mechanisms (Brubaker et al., 2019a, 2019b; Brubaker and Lauffenburger, 2020). Others have employed regression-based approaches to analyze large compendia of mouse and human inflammatory gene expression datasets to identify disease-gene associations across species (Normand et al., 2018). In UC, the work by Czarnewski employs an unsupervised learning approach to analyze data from multiple UC cohorts in conjunction with time-series gene expression data from the dextran sodium sulfate (DSS) murine model of colitis to identify therapeutic responsive and resistant subsets of UC patients (Czarnewski et al., 2019). Like the method proposed in Czarnewski et al., TransComp-R is capable of making comparisons across species when phenotypes, time points, and covariates are not concordant between mice and humans, showing the advantage of both cross-species approaches over single species analysis. Overall, there is tremendous opportunity for systems biology approaches to cross-species modeling to improve patient stratification and therapeutic positioning.
In summary, we have integrated transcriptomics of preclinical models and human IBD to predict the efficacy of a novel kinase inhibitor to alleviate intestinal inflammation. Our results show that biological variation between mouse models can predict clinically relevant phenotypic differences between patients and modeling biological information from multiple mouse models may be a valuable tool in translational systems biology going forward.
Limitations of the study
Our efforts of transcriptomic profiling after acute MK2i treatment explored the direct impact of MK2 inhibition on the tissue state. However, the exact mechanisms underlying such changes could be attributed to transcriptional activities, post-transcriptional modulations, cell state transitions, and/or changes in composition of various cell types. Therefore, further effort would be to focus on deconvoluting the mechanism through which MK2 regulate and contribute to the inflammatory state of the intestinal tissues.
The cross-species modeling of MK2i-responsiveness provides a hypothesis of the IBD clinical population that may benefit from MK2i therapy. Future studies in human cell types examining the interaction between MK2 and TNF-α inhibitors are needed to verify this prediction and elucidate the biological mechanisms by which TNF-α therapy resistant patients may respond to MK2 inhibition therapy.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit anti-MK2 | Cell Signaling Technology | Cat# 3041; RRID:AB_330726 |
| Rabbit anti-phospho-MK2 | Cell Signaling Technology | Cat# 12155; RRID:AB_2797831 |
| Rabbit anti-Hsp27 | Cell Signaling Technology | Cat# 2442; RRID:AB_2233273 |
| Rabbit anti-phospho-Hsp27 | Cell Signaling Technology | Cat# 9709; RRID:AB_11217429 |
| Rabbit anti-GAPDH | Cell Signaling Technology | Cat# 5174; RRID:AB_10622025 |
| Anti-CD45-PacBlue | Biolegend | Cat# 103125; RRID:AB_493536 |
| Anti-Ly6C-APC.Cy7 | Biolegend | Cat# 128025; RRID:AB_10643867 |
| Anti-CD11b-PerCp.Cy5.5 | Biolegend | Cat# 101227; RRID:AB_893233 |
| Anti-GR-1-AF700 | Biolegend | Cat# 108421; RRID:AB_493728 |
| Anti-CD45RB-PE | Biolegend | Cat# 103308; RRID:AB_313015 |
| Anti-CD4-FITC | Biolegend | Cat# 116004; RRID:AB_313689 |
| Critical commercial assays | ||
| Dynabeads™ Untouched™Mouse CD4 Cells Kit | Invitrogen | Cat# 11415D |
| Chemicals, peptides, and recombinant proteins | ||
| MK2 inhibitor ATI-450 | Aclaris | N/A |
| Deposited data | ||
| Bulk RNA-Seq (mouse) | This paper | GEO: GSE164339 |
| Single-cell RNA-Seq (mouse) | Biton et al. (2018) | GEO: GSE106510 |
| Single-cell RNA-Seq (human) | Martin et al., 2019 | GEO: GSE134809 |
| IBD Transcriptomics Data (human) | Arijs et al. (2009) | GEO: GSE16879 |
| Experimental models: Organisms/strains | ||
| Mouse: Rag1-/- | Jackson Laboratories | 002216 |
| Mouse: TNFΔARE | ||
| Mouse: SAMP1/YitFc | Jackson Laboratories | 009355 |
| Mouse: AKR | Jackson Laboratories | 000648 |
| Software and algorithms | ||
| FastQC (0.11.5) | Andrews S. (2010). FastQC: a quality control tool for high throughput sequence data. | https://www.bioinformatics.babraham.ac.uk/projects/fastqc/ |
| Qualimap (2.2.2) | García-Alcalde et al. (2012) | http://qualimap.conesalab.org |
| Salmon (1.4.0) | Patro et al. (2017) | https://salmon.readthedocs.io/en/latest/salmon.html |
| Mouse reference genome Ensembl (Mus_musculus.GRCm38) | Yates et al. (2020) | https://useast.ensembl.org/info/data/index.html |
| DESeq2 (1.28.0) | Love et al. (2014) | https://bioconductor.org/packages/release/bioc/html/DESeq2.html |
| fgsea (1.14.0) | Korotkevich et al. (2016) | https://bioconductor.org/packages/release/bioc/html/fgsea.html |
| clusterProfiler (3.16.1) | Yu et al. (2012) | https://bioconductor.org/packages/release/bioc/html/clusterProfiler.html |
| Seurat (3.1.5) | Stuart et al. (2019) | https://satijalab.org/seurat/articles/install.html |
| GSVA (1.38.2) | Hänzelmann et al. (2013) | https://bioconductor.org/packages/release/bioc/html/GSVA.html |
| TransComp-R | Brubaker et al. (2020) | Mathworks FileExchange 77987 |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Kevin M. Haigis (Kevin_Haigis@dfci.harvard.edu).
Materials availability
This study did not generate new unique reagents.
Experimental model and subject Details
Mice
T cell transfer colitis was induced in Rag1 null mice as described before (Lyons et al., 2018; Strasser et al., 2019). Briefly, splenocytes were purified from 8-weeks old wildtype (WT) C57BL/6 male mice (Jackson Laboratory, Bar Harbor, ME) and subsequently processed for CD4 T cell enrichment using the Dynabeads™ Untouched™ Mouse CD4 Cells Kit, following manufacturer’s instructions. Resulting single cells populations were stained with CD4 and CD45Rb fluorescently labelled antibodies. Naïve T cells were obtained by FACS sorting and defined as the 40% CD45RB brightest population in CD4 positive cells. Bulk T cells as well as controls were sorted and defined as the 15% dimmest CD45RB positive population in CD4 positive gated population.
Naïve and Bulk T cells were injected intraperitoneally in 8-12 weeks old Rag1 null mice at 400,000 and 200,000 cells per animal, respectively. Naïve T injected mice developed colitis in an 8-12 week period after injection, whereas Bulk T injected mice served as uninflamed controls.
Animals were then monitored for colitis with direct screening by rigid colonoscopy. Optical measurements were performed using the Image 1 camera system (Karl-Storz, Tuttlingen, Germany) and UR-4MD HD video recorder (TEAC, Montebello, CA). Once clear symptoms of colitis were developed (weight loss, diarrhea, wall transparency loss and bleeding ulcers), mice were randomized into treatment groups.
TNFΔARE mice were maintained in a heterozygous breeding scheme in order to generate inflamed (Tnf ΔARE/+) and uninflamed littermate controls (Tnf +/+). When mice reached 16 weeks of age, were randomized into treatment groups for MK2 inhibition studies.
SAMP1/YitFc mice and sex and age matched AKR controls were purchased from Jackson Laboratories (Jackson Laboratory, Bar Harbor, ME). At 20 weeks of age, mice were derived for preclinical studies, as clear symptoms of inflammation are present at that age.
Both male and female mice were used in all our studies. All mouse procedures described here were approved by the BIDMC and DFCI Institutional Committees for Animal Care and conducted in compliance with the Animal Welfare Act regulations and other federal statutes relating to animals and experiments involving animals and adhere to the principles set forth in the Guide for the Care and Use of Laboratory Animals, National Research Council, 1996 (Institutional Animal Welfare Assurance No. A-3125–01).
Method details
MK2 inhibitor treatments
MK2 inhibitor ATI-450 was obtained from Aclaris Therapeutics. The drug was formulated in the chow at 3000ppm and given to the mice ad libitum for a total of two (TCT and SAMP1/YitFc mice) or three weeks (TNFΔARE mice). Regular chow was used as vehicle control.
For transcriptomic studies, mice were dosed with 3mg/kg ATI450 or vehicle (0.5% methylcellulose/0.025% Tween20/PBS), and re-dosed 16 hours later. 90 min after the second dose, mice were euthanized and tissues harvested for RNA extraction.
Pathology evaluation
Intestinal tissues were prepared in swiss rolls for pathology evaluation. Briefly, rolls were fixed overnight in neutral buffered formalin and paraffin embedded. 4μm slides were cut and H&E stained. Inflammation scores were blindly assigned under a light microscope. Scoring system was based on (Erben et al., 2014). Briefly, 0 (normal) to 5 (severe) scores were given based on the extent of immune infiltrates (mucosa/submucosa/transmural), presence of epithelial changes (hyperplasia, goblet cells loss, erosions), and mucosal architecture (irregular crypts, crypt loss, villi blunting). Given the diverse kinetics of the different mouse models utilized, we applied slightly different scoring criteria to evaluate the severity of inflammation on each model. All scoring systems ranged from 0 to 5, and mice were randomized into vehicle or treatment groups once they reached full-blown inflammation, which corresponds to a score of 5. Detailed scoring system criteria is depicted in Table S22.
Luminex and western blot
Colon or Ileum tissues were snap frozen and brought to powder in liquid nitrogen with a tissue homogenizer. Tissue homogenates were then lysed in Bioplex buffer containing proteases/phosphatases inhibitors cocktails (BioRad). Multiplex based cytokine/chemokine quantification was performed using the Milliplex MAP Mouse Cytokine/Chemokine magnetic bead panel (Millipore) in a MAGPIX instrument, following manufacturer’s instructions.
For western blotting, 20μg of total protein was resolved by SDS-PAGE before immunoblotting. Primaries antibodies were used as follows: anti-phospho-MK2 (CST 3041, 1:2000), anti-MK2 (CST 12155, 1:2000), anti-phospho-Hsp27 (CST 9709, 1:2000) anti-Hsp27 (CST 2442, 1:2000) anti-GAPDH (CST 8174 1:2000). Fluorescently labelled secondary antibodies were used for protein detection in Odyssey fluorescence imaging system. ImageStudio software was used to quantify band pixel intensity.
FACS analysis of immune populations
Colons or Ileums (after Peyer’s patches removal) were flushed with cold PBS and then washed first in a HBSS containing 5mM EDTA, 1mM DTT, 15mM Hepes and 5% FBS for 30 min at 37°C with agitation. Then, tissue was washed in HBSS, 1mM EDTA, 5% FBS for 10 min at 37°C with agitation and then incubated for 5 min at room temperature in HBSS, 15mM Hepes, 5% FBS to strip the epithelial later. Tissues were then minced with scissors and digested with Liberase TL (0.3mg/ml)/DNAse (10μg/ml) solution for 40 min at 37°C to obtain lymphocyte populations infiltrated in the intestinal mucosa. Digested tissue was then filtered through 100uM mesh and washed in PBS solution containing 1% FBS, 2mM EDTA. Single cell suspensions were incubated for 10 min at room temperature with Fc blocking (CD16/32) antibody (eBioscience) prior to staining with fluorochrome-conjugated antibodies against mixtures of the following antigens: CD45, CD11b, GR1 and Ly6C. Thermo Blue Live/Dead staining was used to exclude dead cells. Stained single cells were fixed with EC fixation kit (eBioscience) and kept at 4°C before FACS analysis. Multiparameter acquisition was performed on a Cytoflex (Beckman Coulter) and bi-dimensional dot plots were generated using FlowJo software.
RNA-seq sequencing
Distal colons and ileum tissues were snap-frozen and powderized in liquid nitrogen with a tissue homogenizer. RNA was extracted using the RNeasy Plus Universal kit from Qiagen, following the manufacturer's instructions. RNA concentration and quality were evaluated in Agilent Bioanalyzer/TapeStation. 1μg of RNA was used for RNA-Seq library preparation. Poly-A enrichment was performed to increase mRNA amount in the input RNA. RNA-Seq library was prepared using NEBNext® Ultra™ II Directional RNA Library Prep Kit for Illumina® following the manufacturer's instructions. Quality control of prepare libraries was performed using RNA TapeStation and qPCR. Libraries were sequenced on Illumina® NextSeq 500 with 10 million reads/sample in 75bp single read mode.
RNA-seq analysis
Quality control of the sequencing result was performed using FastQC (0.11.5) (Andrews S.(2010). FastQC: a quality control tool for high throughput sequence data. Available online at: http://www.bioinformatics.babraham.ac.uk/projects/fastqc) and Qualimap (2.2.2) (García-Alcalde et al., 2012). Salmon (1.4.0) pseudo-aligner was used for sequencing alignment, with the Ensembl mm10 genome (Mus_musculus.GRCm38) as reference (Patro et al., 2017; Yates et al., 2020). Differential gene expression analysis was done using “DESeq2” (1.28.0) with default setting (Love et al., 2014). Genes with expression detected in less than 2 samples in any condition were removed from the analysis. Pre-ranked GSEA analysis was performed using “fgsea” (1.14.0) package with MsigDB Hallmark gene-set using gene expression LFC for ranking (Korotkevich et al., 2016). GO and KEGG analysis was done using the "clusterProfiler” (3.16.1) package (Yu et al., 2012).
Single-cell RNA-Seq analysis
Raw data of the published dataset (GEO: GSE106510& GSE134809) were acquired from the Broad Institute Single Cell Portal (singlecell.broadinstitute.org) or GEO directly. Data quality control, integration, cell clustering, and post-clustering analysis were performed using “Seurat” (3.1.5) package (Stuart et al., 2019). GSVA analysis was performed using the “GSVA” (1.38.2) package (Hänzelmann et al., 2013) with the MsigDB Hallmark gene set. MK2i Signature Score is calculated using method adapted from (Biton et al., 2018). 25 MK2i signature genes were detected in the dataset: Raf1, Akt1, Cdk1, Csnk2b, Mknk1, Myd88, Nfkbib, Arhgdia, Rac1, Ripk1, Stat2, Tnfrsf1a, Vav2, Ywhaz, Pdk1, Trib3, Dapp1, Map2k3, Mapk8, Tbk1, Ikbke, Gsk3a, Actr3, Ap2m1, Ptpn6. To calculate the signature score, a background gene set of 250 (25∗10) genes were selected. These genes were the nearest gene neighbors to the 25 signature genes in the 2-D space defined by mean expression and detection frequency of all cells. The background gene set was used to normalize for the different sequencing depth and library complexity between cells. The signature score was then calculated as the mean expression of the 25 signature genes minus the mean expression of the 250 background genes in each cell.
Cross-species translation modeling
Gene expression data for IBD subtypes from Ulcerative Colitis (UC), colonic and ileal Crohn’s disease (cCD and iCD) patients was obtained from Gene Expression Omnibus (GEO) using the robust multichip average method and Bioconductor tools (Biobase 2.30.0, GEOquery 2.40.0, limma 3.26.8) (GEO Accession: GSE16879) (Arijs et al., 2009; Edgar et al., 2002; Gentleman et al., 2004; Irizarry et al., 2003). For cross-species modeling, all mouse genes with one-to-one human homologs were converted to human gene identifiers using the mouse genome informatics (MGI) database (Blake et al., 2017; Eppig et al., 2015). We identified genes differentially expressed between MK2i responsive mice (TCT) and non-responsive mice (TNFΔARE) (Wilcoxon Mann-Whitney, False Discovery Rate q< 0.05). These genes were used to train a human principal component analysis (PCA) model for each disease subtype and a generalized linear model (GLM) was employed to identify human PC’s predictive of disease status and response to Infliximab. We employed Translatable Components Regression (TransComp-R, MATLAB_R2020b) to identify human PC’s predictive of mouse model type as a proxy for sensitivity to MK2 inhibition (TCT or TNFΔARE) and mouse inflammation status (Brubaker et al., 2020). Gene loading coefficients on translatable human PC’s were evaluated by GSEA PreRanked (v.4.1) (Subramanian et al., 2005).
Prediction of SAMP1/YitFc response to MK2i
Intestinal gene expression data for SAMP1/YitFc mice (SAMP) was obtained from GEO and converted to human gene symbols using the MGI database (GSE124825) (Blake et al., 2000; Edgar et al., 2002; Eppig et al., 2015). SAMP1/YitFc mouse samples were then projected onto human iCD PC'’s and hierarchical clustering was employed to assess whether the SAMP mice projected more similarly to TCT or TNFΔARE mice, an indicator of the likelihood of this mouse model to respond to MK2 inhibition.
Quantification and statistical analysis
Unless stated otherwise, statistical analysis was performed using GraphPad Prism software 9 (GraphPad Software, San Diego, CA). Data are presented as the mean ± SEM unless indicated otherwise. Comparisons of two unpaired groups were calculated using unpaired two-tailed Student’s t-tests, while comparisons of paired samples were calculated using Wilcoxon rank sum test. Comparisons of more than two groups were calculated using two-way analysis of variance (ANOVA) with post hoc Tukey’s multiple comparisons test. P values and statistical analysis are indicated in the figure legends.
Acknowledgments
We are grateful for the provision of mice from Ken Lau at Vanderbilt University. This work was supported by grants from the U.S. Department of Defense (W81XWH-16-1-0042 to K.M.H. and W911NF-19-2-0026 to D.A.L), the National Institutes of Health (U01-CA215798 to D.A.L. and K.M.H.), and a sponsored research agreement from Aclaris Pharmaceuticals. L.S-L. was supported by a postdoctoral fellowship from the Crohn's and Colitis Foundation of America.
Author contributions
Conceptualization, L.S-L.and K.H.; Methodology, L.S-L., B.S. and D.B.; Software, B.S. and D.B.; Validation, L.S-L., B.S. and M.H.; Formal analysis, L.S-L, B.S. and D.B.; Investigation, L.S-L., M.H., A.L. and A.S.; Resources, P.C. and D.L.; Writing L.S-L., B.S., D.B. and K.H.; Funding acquisition, D.L and K.H.; Supervision, K.H.
Declaration of interests
P.C. is an employee of Aclaris Pharmaceuticals. D.K.B. is a paid consultant for Boehringer Ingelheim Pharmaceuticals. K.M.H. is a paid consultant for Aclaris Pharmaceuticals.
Inclusion and diversity
We worked to ensure sex balance in the selection of non-human subjects.
Published: December 17, 2021
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2021.103406.
Supplemental information
Data and code availability
-
•
All mouse RNA-Seq data were deposited in NCBI Gene Expression Omnibus (GEO) with accession number GSE164339 and is publicly available as of the date of publication. Publicly available scRNA-Seq data used in this study are available from GEO under accession GSE106510 (mouse) and GSE134809 (human) and DOI are listed in the key resources table.
-
•
Code used for mouse RNA-Seq analysis and scRNA-Seq analysis are available at https://github.com/sbdaxia/RNASeq_IBD. and is publicly available as of the date of publication. DOIs are listed in the key resources table. The cross-species modeling code for TransComp-R is available on Mathworks FileExchange ID:77987.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- Arijs I., De Hertogh G., Lemaire K., Quintens R., Van Lommel L., Van Steen K., Leemans P., Cleynen I., Van Assche G., Vermeire S., et al. Mucosal gene expression of antimicrobial peptides in inflammatory bowel disease before and after first infliximab treatment. PLoS One. 2009;4 doi: 10.1371/journal.pone.0007984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biton M., Haber A.L., Rogel N., Burgin G., Beyaz S., Schnell A., Ashenberg O., Su C.W., Smillie C., Shekhar K., et al. T helper cell cytokines modulate intestinal stem cell renewal and differentiation. Cell. 2018;175:1307–1320.e22. doi: 10.1016/j.cell.2018.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blake J.A., Eppig J.T., Kadin J.A., Richardson J.E., Smith C.L., Bult C.J., Anagnostopoulos A., Baldarelli R.M., Beal J.S., Bello S.M., et al. Mouse Genome Database (MGD)-2017: community knowledge resource for the laboratory mouse. Nucleic Acids Res. 2017;45:D723–D729. doi: 10.1093/nar/gkw1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blake P.S., Taylor D.R., Crisp C.M., Mander L.N., Owen D.J. Identification of endogenous gibberellins in strawberry, including the novel gibberellins GA123, GA124 and GA125. Phytochemistry. 2000;55:887–890. doi: 10.1016/s0031-9422(00)00237-5. [DOI] [PubMed] [Google Scholar]
- Brubaker D.K., Kumar M.P., Chiswick E.L., Gregg C., Starchenko A., Vega P.N., Southard-Smith A.N., Simmons A.J., Scoville E.A., Coburn L.A., et al. An interspecies translation model implicates integrin signaling in infliximab-resistant inflammatory bowel disease. Sci. Signal. 2020;13 doi: 10.1126/scisignal.aay3258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brubaker D.K., Lauffenburger D.A. Translating preclinical models to humans. Science. 2020;367:742–743. doi: 10.1126/science.aay8086. [DOI] [PubMed] [Google Scholar]
- Brubaker D.K., Paulo J.A., Sheth S., Poulin E.J., Popow O., Joughin B.A., Strasser S.D., Starchenko A., Gygi S.P., Lauffenburger D.A., Haigis K.M. Proteogenomic network analysis of context-specific KRAS signaling in mouse-to-human cross-species translation. Cell Syst. 2019;9:258–270.e6. doi: 10.1016/j.cels.2019.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brubaker D.K., Proctor E.A., Haigis K.M., Lauffenburger D.A. Computational translation of genomic responses from experimental model systems to humans. PLOS Comput.Biol. 2019;15 doi: 10.1371/journal.pcbi.1006286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buttó L.F., Pelletier A., More S.K., Zhao N., Osme A., Hager C.L., Ghannoum M.A., Sekaly R.P., Cominelli F., Dave M. Intestinal stem cell niche defects result in impaired 3D organoid formation in mouse models of Crohn’s disease-like ileitis. Stem Cell Rep. 2020;15:389–407. doi: 10.1016/j.stemcr.2020.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao H., Deterding L.J., Blackshear P.J. Phosphorylation site analysis of the anti-inflammatory and mRNA-destabilizing protein tristetraprolin. Expert Rev. Proteomics. 2007 doi: 10.1586/14789450.4.6.711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czarnewski P., Parigi S.M., Sorini C., Diaz O.E., Das S., Gagliani N., Villablanca E.J. Conserved transcriptomic profile between mouse and human colitis allows unsupervised patient stratification. Nat. Commun. 2019;10 doi: 10.1038/s41467-019-10769-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dambach D.M. Potential adverse effects associated with inhibition of p38alpha/beta MAP kinases. Curr.Top. Med. Chem. 2005;5:929–939. doi: 10.2174/1568026054985911. [DOI] [PubMed] [Google Scholar]
- Denson L.A., Curran M., McGovern D.P.B., Koltun W.A., Duerr R.H., Kim S.C., Sartor R.B., Sylvester F.A., Abraham C., de Zoeten E.F., et al. Challenges in IBD research: precision medicine. Inflamm.Bowel Dis. 2019;25:S31–S39. doi: 10.1093/ibd/izz078. [DOI] [PubMed] [Google Scholar]
- Duraisamy S., Bajpai M., Bughani U., Dastidar S.G., Ray A., Chopra P. MK2 : a novel molecular target for anti-inflammatory therapy. Expert Opin.Ther. Targets. 2008;12:921–936. doi: 10.1517/14728222.12.8.921. [DOI] [PubMed] [Google Scholar]
- Edgar R., Domrachev M., Lash A.E. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002;30:207–210. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engel K., Kotlyarov A., Gaestel M. Leptomycin B-sensitive nuclear export of MAPKAP kinase 2 is regulated by phosphorylation. EMBO J. 1998;17:3363–3371. doi: 10.1093/emboj/17.12.3363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enslen H., Raingeaud J., Davis R.J. Selective activation of p38 mitogen-activated protein (MAP) kinase isoforms by the MAP kinase kinases MKK3 and MKK6. J. Biol. Chem. 1998;273:1741–1748. doi: 10.1074/jbc.273.3.1741. [DOI] [PubMed] [Google Scholar]
- Eppig J.T., Richardson J.E., Kadin J.A., Ringwald M., Blake J.A., Bult C.J. Mouse genome informatics (MGI): reflecting on 25 years. Mamm. Genome. 2015;26:272–284. doi: 10.1007/s00335-015-9589-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erben U., Loddenkemper C., Doerfel K., Spieckermann S., Haller D., Heimesaat M.M., Zeitz M., Siegmund B., Kühl A.A. A guide to histomorphological evaluation of intestinal inflammation in mouse models. Int. J. Clin. Exp. Pathol. 2014;7:4557–4576. [PMC free article] [PubMed] [Google Scholar]
- Feagan B.G., Rutgeerts P., Sands B.E., Hanauer S., Colombel J.F., Sandborn W.J., Van Assche G., Axler J., Kim H.J., Danese S., et al. Vedolizumab as induction and maintenance therapy for ulcerative colitis. N. Engl. J. Med. 2013;369:699–710. doi: 10.1056/NEJMoa1215734. [DOI] [PubMed] [Google Scholar]
- Fiore M., Forli S., Manetti F. Targeting mitogen-activated protein kinase-activated protein kinase 2 (MAPKAPK2, MK2): medicinal chemistry efforts to lead small molecule inhibitors to clinical trials. J. Med. Chem. 2016;59:3609–3634. doi: 10.1021/acs.jmedchem.5b01457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaestel M., Kotlyarov A., Kracht M. Targeting innate immunity protein kinase signalling in inflammation. Nat. Rev. Drug Discov. 2009;8:480–499. doi: 10.1038/nrd2829. [DOI] [PubMed] [Google Scholar]
- García-Alcalde F., Okonechnikov K., Carbonell J., Cruz L.M., Götz S., Tarazona S., Dopazo J., Meyer T.F., Conesa A. Qualimap: evaluating next-generation sequencing alignment data. Bioinformatics. 2012;28:2678–2679. doi: 10.1093/bioinformatics/bts503. [DOI] [PubMed] [Google Scholar]
- Gentleman R.C., Carey V.J., Bates D.M., Bolstad B., Dettling M., Dudoit S., Ellis B., Gautier L., Ge Y., Gentry J., et al. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5 doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hänzelmann S., Castelo R., Guinney J. GSVA: gene set variation analysis for microarray and RNA-Seq data. BMC Bioinform. 2013;14 doi: 10.1186/1471-2105-14-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irizarry R.A., Hobbs B., Collin F., Beazer-Barclay Y.D., Antonellis K.J., Scherf U., Speed T.P. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4:249–264. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- Johnson B.A., Stehn J.R., Yaffe M.B., Keith Blackwell T. Cytoplasmic localization of tristetraprolin involves 14-3-3-dependent and -independent mechanisms. J. Biol. Chem. 2002;277:18029–18036. doi: 10.1074/jbc.M110465200. [DOI] [PubMed] [Google Scholar]
- Kontoyiannis D., Boulougouris G., Manoloukos M., Armaka M., Apostolaki M., Pizarro T., Kotlyarov A., Forster I., Flavell R., Gaestel M., et al. Genetic dissection of the cellular pathways and signaling mechanisms in modeled tumor necrosis factor–induced crohn’s-like inflammatory bowel disease. J. Exp. Med. 2002;196:1563–1574. doi: 10.1084/jem.20020281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kontoyiannis D., Pasparakis M., Pizarro T.T., Cominelli F., Kollias G. Impaired on/off regulation of TNF biosynthesis in mice lacking TNF AU- rich elements: implications for joint and gut-associated immunopathologies. Immunity. 1999;10:387–398. doi: 10.1016/S1074-7613(00)80038-2. [DOI] [PubMed] [Google Scholar]
- Korotkevich G., Sukhov V., Sergushichev A. Fast gene set enrichment analysis. bioRxiv. 2016:060012. doi: 10.1101/060012. [DOI] [Google Scholar]
- Kotlyarov A., Neininger A., Schubert C., Eckert R., Birchmeier C., Volk H.D., Gaestel M. MAPKAP kinase 2 is essential for LPS-induced TNF-alpha biosynthesis. Nat. Cell Biol. 1999;1:94–97. doi: 10.1038/10061. [DOI] [PubMed] [Google Scholar]
- Li Y.Y., Yuece B., Cao H.M., Lin H.X., Lv S., Chen J.C., Ochs S., Sibaev A., Deindl E., Schaefer C., Storr M. Inhibition of p38/Mk2 signaling pathway improves the anti-inflammatory effect of WIN55 on mouse experimental colitis. Lab. Investig. 2013;93:322–333. doi: 10.1038/labinvest.2012.177. [DOI] [PubMed] [Google Scholar]
- Limbourg A., von Felden J., Jagavelu K., Krishnasamy K., Napp L.C., Kapopara P.R., Gaestel M., Schieffer B., Bauersachs J., Limbourg F.P., et al. MAP-kinase activated protein kinase 2 links endothelial activation and monocyte/macrophage recruitment in arteriogenesis. PLoS One. 2015;10:1–12. doi: 10.1371/journal.pone.0138542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15 doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowell C.A., Varmus H.E. Functional overlap in the src gene family: inactivation of hck and fgr impairs natural immunity. Genes Dev. 1994;8:387–398. doi: 10.1101/gad.8.4.387. [DOI] [PubMed] [Google Scholar]
- Lyons J., Ghazi P.C., Starchenko A., Tovaglieri A., Baldwin K.R., Poulin E.J., Gierut J.J., Genetti C., Yajnik V., Breault D.T., et al. The colonic epithelium plays an active role in promoting colitis by shaping the tissue cytokine profile. PLoS Biol. 2018;16:1–24. doi: 10.1371/journal.pbio.2002417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin J.C., Chang C., Boschetti G., Ungaro R., Giri M., Grout J.A., Gettler K., Chuang L., Nayar S., Greenstein A.J., et al. Single-cell analysis of Crohn’s disease lesions identifies a pathogenic cellular module associated with resistance to Anti-TNF therapy. Cell. 2019;178:1493–1508. doi: 10.1016/j.cell.2019.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto S., Okabe Y., Setoyama H., Takayama K., Ohtsuka J., Funahashi H., Imaoka A., Okada Y., Umesaki Y. Inflammatory bowel disease-like enteritis and caecitis in a senescence accelerated mouse P1/Yit strain. Gut. 1998;43:71–78. doi: 10.1136/gut.43.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen O.H., Ainsworth M.A. Tumor necrosis factor inhibitors for inflammatory bowel disease. N. Engl. J. Med. 2013;369:754–762. doi: 10.1056/NEJMct1209614. [DOI] [PubMed] [Google Scholar]
- Normand R., Du W., Briller M., Gaujoux R., Starosvetsky E., Ziv-Kenet A., Shalev-Malul G., Tibshirani R.J., Shen-Orr S.S. Found in translation: a machine learning model for mouse-to-human inference. Nat. Methods. 2018;15:1067–1073. doi: 10.1038/s41592-018-0214-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papamichael K., Gils A., Rutgeerts P., Levesque B.G., Vermeire S., Sandborn W.J., Vande Casteele N. Role for therapeutic drug monitoring during induction therapy with TNF antagonists in IBD: evolution in the definition and management of primary nonresponse. Inflamm. Bowel Dis. 2015 doi: 10.1097/MIB.0000000000000202. [DOI] [PubMed] [Google Scholar]
- Patro R., Duggal G., Love M.I., Irizarry R.A., Kingsford C. Salmon provides fast and bias-aware quantification of transcript expression. Nat. Methods. 2017;14:417–419. doi: 10.1038/nmeth.4197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pizarro T.T., Arseneau K.O., Bamias G., Cominelli F. Mouse models for the study of Crohn’s disease. Trends Mol. Med. 2003 doi: 10.1016/S1471-4914(03)00052-2. [DOI] [PubMed] [Google Scholar]
- Pizarro T.T., Pastorelli L., Bamias G., Garg R.R., Reuter B.K., Mercado J.R., Chieppa M., Arseneau K.O., Ley K., Cominelli F. SAMP1/YitFc mouse strain: a spontaneous model of Crohn’s disease-like ileitis. Inflamm.Bowel Dis. 2011;17:2566–2584. doi: 10.1002/ibd.21638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandborn W.J., Rutgeerts P., Feagan B.G., Reinisch W., Olson A., Johanns J., Lu J., Horgan K., Rachmilewitz D., Hanauer S.B., et al. Colectomy rate comparison after treatment of ulcerative colitis with placebo or infliximab. Gastroenterology. 2009;137:1250–1260. doi: 10.1053/j.gastro.2009.06.061. [DOI] [PubMed] [Google Scholar]
- Sands B.E., Peyrin-Biroulet L., Loftus E.V., Danese S., Colombel J.F., Törüner M., Jonaitis L., Abhyankar B., Chen J., Rogers R., et al. Vedolizumab versus Adalimumab for moderate-to-severe ulcerative colitis. N. Engl. J. Med. 2019;381:1215–1226. doi: 10.1056/NEJMoa1905725. [DOI] [PubMed] [Google Scholar]
- Sands B.E., Sandborn W.J., Panaccione R., O’Brien C.D., Zhang H., Johanns J., Adedokun O.J., Li K., Peyrin-Biroulet L., Van Assche G., et al. Ustekinumab as induction and maintenance therapy for ulcerative colitis. N. Engl. J. Med. 2019;381:1201–1214. doi: 10.1056/NEJMoa1900750. [DOI] [PubMed] [Google Scholar]
- Segal B.H., Kuhns D.B., Ding L., Gallin J.I., Holland S.M. Thioglycollate peritonitis in mice lacking C5, 5-lipoxygenase, or p47(phox): complement, leukotrienes, and reactive oxidants in acute inflammation. J. Leukoc Biol. 2002;71:410–416. [PubMed] [Google Scholar]
- Singh S., Pardi D.S. Update on anti-tumor necrosis factor Agents in Crohn disease. Gastroenterol.Clin.North Am. 2014 doi: 10.1016/j.gtc.2014.05.008. [DOI] [PubMed] [Google Scholar]
- Strasser S.D., Ghazi P.C., Starchenko A., Boukhali M., Edwards A., Suarez-Lopez L., Lyons J., Changelian P.S., Monahan J.B., Jacobsen J., et al. Substrate-based kinase activity inference identifies MK2 as driver of colitis. Integr.Biol. (Camb). 2019;11:301–314. doi: 10.1093/intbio/zyz025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuart T., Butler A., Hoffman P., Hafemeister C., Papalexi E., Mauck W.M., Hao Y., Stoeckius M., Smibert P., Satija R. Comprehensive integration of single-cell data. Cell. 2019;177:1888–1902.e21. doi: 10.1016/j.cell.2019.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suarez-Lopez L., Sriram G., Kong Y.W.Y.W., Morandell S., Merrick K.A.K.A., Hernandez Y., Haigis K.M.K.M., Yaffe M.B.M.B. MK2 contributes to tumor progression by promoting M2 macrophage polarization and tumor angiogenesis. Proc. Natl. Acad. Sci. U. S. A. 2018;115:E4236–E4244. doi: 10.1073/pnas.1722020115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian A., Tamayo P., Mootha V.K., Mukherjee S., Ebert B.L., Gillette M.A., Paulovich A., Pomeroy S.L., Golub T.R., Lander E.S., Mesirov J.P. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. U. S. A. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun L., Wu Q., Nie Y., Cheng N., Wang R., Wang G., Zhang D., He H., Ye R.D., Qian F. A role for MK2 in enhancing neutrophil-derived ROS production and aggravating liver ischemia/reperfusion injury. Front. Immunol. 2018;9:2610. doi: 10.3389/fimmu.2018.02610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ter Haar E., Prabakhar P., Liu X., Lepre C. Crystal structure of the P38α-MAPKAP kinase 2 heterodimer. J. Biol. Chem. 2007;282:9733–9739. doi: 10.1074/jbc.M611165200. [DOI] [PubMed] [Google Scholar]
- Wang C., Hockerman S., Jacobsen E.J., Alippe Y., Selness S.R., Hope H.R., Hirsch J.L., Mnich S.J., Saabye M.J., Hood W.F., et al. Selective inhibition of the p38α MAPK–MK2 axis inhibits inflammatory cues including inflammasome priming signals. J. Exp. Med. 2018;215:1315–1325. doi: 10.1084/jem.20172063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yates A.D., Achuthan P., Akanni W., Allen James, Allen Jamie, Alvarez-Jarreta J., Amode M.R., Armean I.M., Azov A.G., Bennett R., et al. Ensembl 2020. Nucleic Acids Res. 2020;48:D682–D688. doi: 10.1093/nar/gkz966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu G., Wang L.G., Han Y., He Q.Y. ClusterProfiler: an R package for comparing biological themes among gene clusters. Omi. A. J. Integr. Biol. 2012;16:284–287. doi: 10.1089/omi.2011.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang T., Jiang J., Liu J., Xu L., Duan S., Sun L., Zhao W., Qian F. MK2 is required for neutrophil-derived ROS production and inflammatory bowel disease. Front. Med. 2020;7 doi: 10.3389/fmed.2020.00207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Y., Humphry M., Maguire J.J., Bennett M.R., Clarke M.C.H. Intracellular interleukin-1 receptor 2 binding prevents cleavage and activity of interleukin-1α, controlling necrosis-induced sterile inflammation. Immunity. 2013;38:285–295. doi: 10.1016/j.immuni.2013.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
All mouse RNA-Seq data were deposited in NCBI Gene Expression Omnibus (GEO) with accession number GSE164339 and is publicly available as of the date of publication. Publicly available scRNA-Seq data used in this study are available from GEO under accession GSE106510 (mouse) and GSE134809 (human) and DOI are listed in the key resources table.
-
•
Code used for mouse RNA-Seq analysis and scRNA-Seq analysis are available at https://github.com/sbdaxia/RNASeq_IBD. and is publicly available as of the date of publication. DOIs are listed in the key resources table. The cross-species modeling code for TransComp-R is available on Mathworks FileExchange ID:77987.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.