Abstract
The DNA binding activity of FUS (also known as TLS), a nuclear pro-oncogene involved in multiple translocations, is regulated by BCR-ABL in a protein kinase CβII (PKCβII)-dependent manner. We show here that in normal myeloid progenitor cells FUS, although not visibly ubiquitinated, undergoes proteasome-dependent degradation, whereas in BCR-ABL-expressing cells, degradation is suppressed by PKCβII phosphorylation. Replacement of serine 256 with the phosphomimetic aspartic acid prevents proteasome-dependent proteolysis of FUS, while the serine-256-to-alanine FUS mutant is unstable and susceptible to degradation. Ectopic expression of the phosphomimetic S256D FUS mutant in granulocyte colony-stimulating factor-treated 32Dcl3 cells induces massive apoptosis and inhibits the differentiation of the cells escaping cell death, while the degradation-prone S256A mutant has no effect on either survival or differentiation. FUS proteolysis is induced by c-Jun, is suppressed by BCR-ABL or Jun kinase 1, and does not depend on c-Jun transactivation potential, ubiquitination, or its interaction with Jun kinase 1. In addition, c-Jun-induced FUS proteasome-dependent degradation is enhanced by heterogeneous nuclear ribonucleoprotein (hnRNP) A1 and depends on the formation of a FUS-Jun-hnRNP A1-containing complex and on lack of PKCβII phosphorylation at serine 256 but not on FUS ubiquitination. Thus, novel mechanisms appear to be involved in the degradation of FUS in normal myeloid cells; moreover, the ability of the BCR-ABL oncoprotein to suppress FUS degradation by the induction of posttranslational modifications might contribute to the phenotype of BCR-ABL-expressing hematopoietic cells.
FUS, also known as TLS or heterogeneous nuclear ribonucleoprotein (hnRNP) P2, was first discovered as the N-terminal part of a fusion with CHOP in myxoid liposarcoma carrying the t(12;16) translocation (8, 33) and was subsequently detected in different types of human myeloid leukemia (37), in which the C terminus of FUS is replaced by the DNA-binding domain of ERG (28). The C terminus of FUS is required for binding to pre-mRNA and mRNA, while the N terminus appears to function as a transcription activation domain (34). FUS is expressed at high levels in hematopoietic and nonhematopoietic tissues and is localized primarily in the nucleus, where it might be involved in pre-mRNA processing and nucleocytoplasmic shuttling, as well as in the regulation of basal transcription (35). FUS expression and DNA binding activity is induced in hematopoietic cells by BCR-ABL (30), which circumvents signals generated by the interaction of growth factors (e.g., interleukin-3 [IL-3]) with their receptors (43). The DNA binding activity of FUS requires protein kinase CβII (PKCβII)-dependent phosphorylation, as indicated by use of PKCβII-specific inhibitors and expression of a dominant-negative PKCβII mutant (30). Suppression of FUS expression in myeloid precursor cells accelerates granulocyte colony-stimulating factor (G-CSF)-stimulated differentiation and is accompanied by upregulation of G-CSF receptor expression (30). By contrast, downregulation of FUS expression in BCR-ABL-expressing cells is associated with suppression of growth factor-independent colony formation, partial restoration of G-CSF-induced granulocytic differentiation, and reduced tumorigenic potential in vivo (30).
The ability of BCR-ABL oncoproteins to transform hematopoietic cells depends on their tyrosine kinase activity (23), which is essential for recruiting and activating multiple biochemical pathways that transduce oncogenic signals (7), positively or negatively regulating the activity of nuclear effectors. The BCR-ABL-dependent activation of nuclear regulators might be due to mechanisms of enhanced transcription, as reported for c-myc (36, 41), but might also involve posttranslational modifications that increase the stability or induce the proteolytic degradation of target substrates. Indeed, BCR-ABL promotes the ubiquitin- and proteasome-dependent degradation of antioncogenic Abi proteins (10).
Oncogenic ABL proteins regulate the activity of many downstream effectors directly or via a cascade of phosphorylation and dephosphorylation events (43). These processes control the formation of multiprotein complexes which appear to be required for transducing oncogenic signals and, perhaps, for regulating the stability of some ABL effectors. Indeed, phosphorylation plays a key role in controlling the function of regulatory proteins by targeting them to the ubiquitin-proteasome proteolytic machinery (13).
In mammalian cells, the 26S proteasome is a specialized multisubunit enzyme with different catalytic activities (22). It is the predominant intracellular, nuclear, and cytoplasmic (3) nonlysosomal proteolytic mechanism involved in the regulation of a broad range of processes, such as cell cycle progression, antigen presentation, and gene expression (6). This degradation pathway involves an enzymatic cascade through which multiple ubiquitin molecules are covalently ligated to the protein substrate, which is then degraded by the 26S proteasome complex (6).
Beside polyubiquitinated substrates, the proteasome is also responsible for the degradation of proteins which, like ornithine decarboxylase (ODC), do not undergo ubiquitination (1, 31).
In this study, we show that FUS is degraded by a proteasome-dependent process in which the targeting of FUS to the proteasome is dependent on the formation of a complex with c-Jun and hnRNP A1 but not on FUS ubiquitination. In BCR-ABL-expressing cells, the enhanced FUS expression requires PKCβII phosphorylation of serine 256, which prevents proteasome-mediated FUS degradation. In parental 32Dcl3 cells treated with G-CSF, ectopic expression of the degradation-resistant serine-to-aspartic acid phosphomimetic FUS mutant induces massive apoptosis and the emergence of a cohort of differentiation-arrested cells able to grow in the presence of G-CSF.
MATERIALS AND METHODS
Cell cultures.
The murine IL-3-dependent 32Dcl3 myeloid precursor and its derivative cell lines were maintained in culture or induced to differentiate as described previously (30). Morphologic differentiation was monitored by May-Grunwald and Giemsa staining of cytospin preparations. For assays requiring cell starvation, cells were washed four times in phosphate-buffered saline (PBS) and incubated for 8 to 12 h in RPMI supplemented with 10% fetal bovine serum or 0.1% bovine serum albumin and 2 mM l-glutamine, as indicated. The 293T human embryonic kidney cell line transformed with the adenovirus 5 DNA (American Type Culture Collection, Rockville, Md.) was maintained in Dulbecco's modified Eagle's medium supplemented with 10% heat-inactivated fetal calf serum and 2 mM glutamine (Gibco). BOSC 23 packaging cells (American Type Culture Collection) were cultured and transfected as described previously (29). The 32DBCR-ABL cell line has been described previously (30).
Transfection and retroviral infection.
32Dcl3 and 32DBCR-ABL-derived cell lines were generated by electroporation (GenPulser; Bio-Rad) at 200 mV and 960 μF with the following retroviral constructs: LXSP-HA-FUS (32D-WTFUS and 32DBCR-ABL-WTFUS), LXSP-HA-S256AFUS (32D-S256AFUS and 32DBCR-ABL-S256AFUS), and LXSP-HA-S256DFUS (32D-S256DFUS). Mixed populations and single-cell clones, obtained after puromycin (2 μg/ml) selection, were maintained in culture as described previously (30). 32Dcl3 and 32DBCR-ABL cells transfected with the empty vector LXSP were morphologically identical to the parental cells. Retroviral infection of parental and BCR-ABL-expressing 32Dcl3 cells was carried out as described previously (29). For transient transfection, 293T cells were grown for 16 to 18 h to 80% confluence and transfected with 30 μg of plasmid DNA by calcium phosphate precipitation using the ProFection system (Promega). The empty pMT plasmid was used to normalize for equal amounts of transfected DNA.
Plasmids. (i) LXSP-HA WT FUS.
A SpeI DNA fragment encoding the hemagglutinin (HA) epitope was subcloned in frame in front of the FUS translation start site after SpeI restriction digestion of plasmid pBS-FUS (30). The resulting plasmid, pSK-HA-FUS, was digested with HindIII and XbaI, Klenow blunt ended, and subcloned in the sense orientation into the blunted EcoRI site of the LXSP retroviral vector.
(ii) LXSP-HA S256A FUS and LXSP-HA S256D FUS.
Primers containing the mutation of FUS serine 256 to alanine or aspartic acid were used to mutagenize FUS (Quickeasy mutagenesis kit; Stratagene) with plasmid pSK-HA-FUS as template. Plasmids pSK-HA S256A FUS and pSK-HA S256D FUS were XbaI-HindIII digested, blunted, and subcloned into the blunted EcoRI site of the LXSP retroviral vector.
(iii) pMT-HA WT FUS.
The XbaI blunted-HindIII fragment from pSK-HA-FUS containing the HA-tagged FUS full-length cDNA was subcloned in the sense orientation into the cytomegalovirus (CMV)-based pMT expression vector (40) previously digested with BamHI, blunted, and digested with HindIII.
(iv) pMT-AZ (antizyme).
The XbaI blunted-HindIII fragment from plasmid ZZ5 (24) containing the full-length antizyme rat cDNA was subcloned in sense orientation into the CMV-based expression vector pMT previously digested with BamHI, blunted, and redigested with HindIII.
(v) pMT-HA-cMyb.
The HA-tagged human c-myb cDNA was subcloned in sense orientation into the CMV-based vector pMT.
(vi) pMT-HA-hnRNP A1.
The full-length hnRNP A1 cDNA (kind gift of G. Dreyfuss, Howard Hughes Medical Institute, University of Pennsylvania School of Medicine, Philadelphia, Pa.) was PCR amplified using an upstream primer containing a BamHI site and a downstream primer containing a mutated stop codon followed by the HA epitope sequence and a HindIII restriction site. The PCR product was BamHI-HindIII digested and subcloned into the CMV-based vector pMT.
(vii) MSCV-6×His-cJun.
The BamHI-HindIII Klenow-blunted fragment containing the His6-tagged c-Jun cDNA was subcloned into the HpaI site of the retroviral vector MSCV-puro (Clontech).
(viii) pGEX-FUS-Pep1(241–270), pGEX-FUS-Pep2(308–337), pGEX-FUS-Pep3(342–376), and pGEX-FUS-Pep4(428–456).
FUS cDNA fragments encoding FUS amino acids 241 to 270 (Pep1), 308 to 337 (Pep2), 342 to 376 (Pep3), and 428 to 456 (Pep4) were generated by PCR performed on plasmid pBS-FUS using upstream primers containing a BamHI restriction site followed by an ATG codon at the 5′ end and downstream primers carrying a stop codon followed by an EcoRI site at the 3′ end. The gel-purified fragments were phosphorylated, digested with BamHI-EcoRI, and directionally subcloned into the BamHI-EcoRI sites of the pGEX-2T vector (Pharmacia Biotech). pGEX-FUS(1–240) and pGEX-FUS(240–526) have been described previously (30). pMT107 (His6-tagged ubiquitin), pMT108 (HA-tagged c-Jun), and pMT35 (His6-tagged c-Jun) were the kind gifts of Dirk Bohmann (European Molecular Biology Laboratories, Heidelberg, Germany). Plasmid ZZ5 was a kind gift from S. Matsufuji (Jikei University, Tokyo, Japan). The LXSP retroviral vector was the kind gift of A. Sacchi (Regina Elena Cancer Institute, Rome, Italy). Wild-type (WT) and dominant negative (APF) Jun NH2-terminal kinase CMV-based expression plasmids (15) were the kind gift of R. J. Davis (University of Massachusetts Medical School, Worcester, Mass.). pRSV-v-Jun was a kind gift of E. J. Black and D. A. F. Gillespie (Beatson Institute for Cancer Research, Bearsden, Glasgow, United Kingdom). The transactivation-deficient S63/73L c-Jun mutant (32) cloned into the mammalian expression vector pMT2 was a kind gift of J. Woodgett (Ontario Cancer Institute, Toronto, Ontario, Canada).
Enzyme inhibitors.
Where indicated, cells were IL-3 starved or IL-3 and serum starved (8 h) in the presence of kinase, protease, or proteasome inhibitors used at the following concentrations: calphostin C, 200 ng/ml (Calbiochem); N-acetyl-Leu-Lev-Nle-CHO (ALLM), 25 μM (Calbiochem); N-acetyl-Leu-Lev-Met-CHO (ALLN), 25 μM (Calbiochem); lactacystin, 10 μM (Calbiochem); and MG 132, 40 μM (Calbiochem).
Western blotting, immunoprecipitation, and Ni-NTA-mediated nickel chromatography.
Cells were harvested, washed twice with ice-cold PBS, and lysed (107 cells/100 μl of lysis buffer) in HEPES buffer (10 mM HEPES [pH 7.5], 150 mM NaCl, 10% [vol/vol] glycerol, 1 mM EDTA, 1 mM dithiothreitol, 1 mM phenylmethylsulfonyl fluoride, 25 μg of aprotinin per ml, 10 μg of leupeptin per ml, 100 μg of pepstatin A per ml, 5 mM benzamidine, 1 mM Na3VO4, 50 mM NaF, 10 mM β-glycerolphosphate) containing 1% (vol/vol) NP-40. Lysates and immunoprecipitated proteins were obtained and processed as described previously (29). Nuclear and cytoplasmic subcellular fractions were obtained as follows. Cells (107) were washed twice in ice-cold PBS and lysed in 1 ml of isotonic buffer (150 mM NaCl, 20 mM HEPES [pH 7.5]) supplemented with 0.2% NP-40. After disruption of the cytoplasmic membrane, nuclei were collected by centrifugation (5 min at 500 × g and 4°C), lysed in isotonic buffer supplemented with 1% NP-40, and clarified by centrifugation. FUS antiserum was obtained by rabbit immunization with the agarose-coupled glutathione S-transferase (GST) FUS(1–240) (N-terminal FUS, amino acids 1 to 240) fusion protein and used at a 1:5,000 dilution. Ni-nitrilotriacetic acid (NTA)-mediated nickel chromatography (Ni-NTA agarose: Qiagen Inc., Valencia, Calif.) was performed under denaturing (4) or nondenaturing conditions (Ni-NTA pull-down assay) as suggested by the manufacturer.
Pulse-chase experiments.
32D WT FUS and 32DBCR-ABL cells expressing WT or S256A HA-tagged FUS were cultured for 90 min in RPMI 1640 without methionine and supplemented with 10% dialyzed FBS (Gibco BRL, Grand Island, N.Y.) and 2 ng of recombinant murine IL-3 (Gibco BRL) per ml at 106 cells/ml. The cells were washed and resuspended (5 × 106 cells/ml) in medium containing 250 μCi of [35S]methionine per ml (NEN; Life Science Products). After 1 h, the cells were washed with methionine-containing RPMI and cultured (105 cells/ml) for 12 h in IL-3-containing medium or in serum- and IL-3-deprived medium, supplemented with an excess of l-methionine (3 mg/ml; Gibco BRL). At different times, the cells were harvested and lysed in isotonic buffer (20 mM HEPES [pH 7.5], 150 mM NaCl, 1% NP-40) supplemented with protease and phosphatase inhibitors used at the indicated concentrations. Precleared extracts were incubated at 4°C for 2 h with protein G plus (Oncogene Research Products)-coupled anti-HA antibody (Babco, Berkeley, Calif.). Immunoprecipitated proteins were resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), visualized by autoradiography upon transfer onto a nitrocellulose membrane, and analyzed by densitometry. The half-lives of WT and S256A FUS proteins (t1/2) were calculated using the formula t1/2 = (0.693t)/ln (Nt/N0) as described previously (24).
In vivo 32P labeling.
WT and S256A FUS-expressing 32DBCR-ABL cells were washed three times in phosphate-free RPMI (Gibco BRL), phosphate purged for 3 h, and incubated for 3 h in phosphate-free RPMI containing 0.3 mCi of [32P]orthophosphate (New England Nuclear, Boston, Mass.) per ml, 0.1% bovine serum albumin, and 25 mM HEPES (pH 7.4). Isotype-matched antibody-precleared and protein G-agarose (Pharmacia, Piscataway, N.J.)-precleared lysates were used in immunoprecipitation with an anti-HA antibody previously coated with protein G-agarose. Immunoprecipitates were resolved by SDS-PAGE (4 to 15% polyacrylamide), transferred to nitrocellulose filters, and exposed for autoradiography.
Recombinant protein purification and PKC assay.
BL-21 (DH3) cells were transformed with plasmid pGEX-FUS(1–240), pGEX-FUS(240–526), pGEX-FUS-Pep1, pGEX-FUS-Pep2, pGEX-FUS-Pep3, or pGEX-FUS-Pep4, encoding, respectively, GST fused in frame with the FUS N-terminal amino acids 1 to 240, C-terminal amino acids 240 to 526, peptide 1 (amino acids 241 to 270 [RGRGGGRGGRGGMGGSDRGGFMKFGGPRDQ]), peptide 2 (amino acids 308 to 337 [GIIKTNKKTGQPMINLYTDRETGKLKGEAT]), peptide 3 (amino acids 342 to 376 [DPPSAKAAIDWFDGKEFSGNPIKVSFATRRADFNR]), or peptide 4 (amino acids 428 to 456 [PNPTCENMNFSWRNECNQCKAPKPDGPGG]) containing putative PKCβII phosphorylation sites (underlined). Purified proteins were obtained as specified by the manufacturer (Pharmacia Biotech).
PKCβII serine/threonine kinase activity was assayed using an in vitro PKC assay kit from Upstate Biotechnology, Inc. (UBI), with a recombinant PKCβII (UBI) and 1 μg of N-terminal GST-FUS(1–240), C-terminal GST-FUS(240–526), GST-FUS-Pep1, GST-FUS-Pep2, GST-FUS-Pep3, or GST-FUS-Pep4 as the substrate, as suggested by the manufacturer (UBI). The reaction products were fractionated by SDS-PAGE (4 to 15% polyacrylamide), and the gel was stained with Coomassie blue, dried, and exposed for autoradiography.
Northern blot analysis.
Total RNA was extracted using Tri-Reagent (Molecular Research Center, Inc.). For Northern blot analysis, RNA (15 μg) was fractionated onto denaturing 1% agarose–6.6% formaldehyde gels, transferred to a nylon membrane (Amersham), and hybridized to radiolabeled full-length FUS cDNA (8).
RESULTS
BCR-ABL prevents proteasome-mediated FUS degradation.
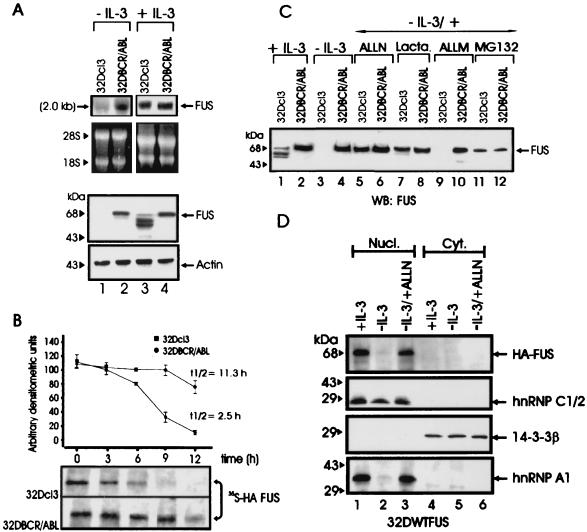
Induction of FUS binding activity in BCR-ABL-expressing 32Dcl3 cells is associated with enhanced expression of FUS (30). To determine whether the induction of FUS expression reflects an increase in mRNA levels or enhanced FUS protein stability, Northern and Western blots were performed using total RNA or cell extracts from parental and BCR-ABL-expressing 32Dcl3 cells cultured in the presence of IL-3 or after 12 h of IL-3 deprivation. Compared to parental 32Dcl3 cells, BCR-ABL-expressing cells showed higher levels of FUS mRNA only when cultured in the absence of IL-3 (Fig. 1A). FUS protein was undetectable in IL-3-starved parental 32Dcl3 cells, whereas low levels of full-length FUS and faster-migrating forms, probably representing FUS degradation products, were detected when these cells where cultured in the presence of IL-3 (Fig. 1A). By contrast, FUS protein was abundant in BCR-ABL-expressing cells regardless of the culture conditions (Fig. 1A). Together, these findings suggest a role of BCR-ABL in preventing FUS degradation. Indeed, pulse-chase experiments performed with parental and BCR-ABL cells grown in the presence of IL-3 and ectopically expressing the HA-tagged FUS to facilitate its detection revealed that the t1/2 of newly synthesized FUS was at least ∼4.5 times longer in BCR-ABL-expressing cells (t1/2 ≈ 11.3 h) than in the parental 32Dcl3 cells (t1/2 ≈ 2.5 h) (Fig. 1B).
FIG. 1.
FUS expression, stability, and proteasome-dependent degradation. (A) Northern (top panel) and Western blot (bottom panel) analysis of FUS expression in parental and BCR-ABL-expressing 32Dcl3 cells in the presence of IL-3 (lanes 3 and 4) or after IL-3-deprivation for 8 h (lanes 1 and 2). rRNA and actin levels were used as controls for RNA and protein loading, respectively. (B) Stability of FUS in IL-3-cultured parental and BCR-ABL-expressing 32Dcl3 cells ectopically expressing the HA-tagged WT FUS. The turnover of FUS was monitored by a pulse-chase assay and quantitated by densitometry. Each point on the graph represents the mean and standard deviation of the relative amount of FUS during the chase period; t1/2 values were calculated using the formula reported in Materials and Methods. The graph is representative of three independent experiments with similar results. (C) Effect of proteasome (lactacystin [Lacta.] and MG132), calpain and proteasome (ALLN), and calpain (ALLM) inhibitors on endogenous FUS expression in IL-3-deprived (8 h), parental, and BCR-ABL-expressing cells. FUS was detected using antiserum raised against the N-terminal region (amino acids 1 to 240) of FUS. (D) Effect of ALLN on nuclear and cytoplasmic levels of HA-tagged FUS. Western blots show expression of HA-tagged FUS, hnRNP C1/2, 14-3-3β, and hnRNP A1 in nuclear and cytoplasmic fractions of 32Dcl3 cells (cultured in IL-3, IL-3 starved [for 8 h], or IL-3 starved in the presence of ALLN). Expression of hnRNP C1/2 was used as nuclear marker, while that of 14-3-3β was used as cytoplasmic marker. The anti-hnRNP C1/2 (4F4) and the anti-hnRNP A1 (9H10) monoclonal antibodies were a kind gift of G. Dreyfuss (Howard Hughes Medical Institute, University of Pennsylvania School of Medicine, Philadelphia, Pa.), while HA-tagged FUS and 14-3-3β were detected using monoclonal anti-HA and anti 14-3-3β (Santa Cruz Biotechnology, Santa Cruz, Calif.) antibodies. Data are representative of three different experiments with similar results.
To investigate which proteolytic pathway might be responsible for FUS degradation, inhibitors of Ca2+-dependent neutral proteases calpains (ALLN and ALLM), caspases (DEVD and ZVAD-FMK), and the proteasome catalytic activities (lactacystin, MG132, and ALLN), were assayed for their ability to rescue FUS expression in 32Dcl3 cells deprived of IL-3 for 8 h. Indeed, FUS expression was restored to levels comparable to those of IL-3-cultured cells only when parental cells were IL-3 starved in the presence of 25 μM ALLN, 40 μM MG132, or 10 μM lactacystin (Fig. 1C), whereas the calpain inhibitor 25 μM ALLM (Fig. 1C) and all the other inhibitors had no effect (data not shown). Thus, FUS degradation appears to be proteasome dependent. Note that FUS levels were not downmodulated by growth factor deprivation of BCR-ABL-expressing cells and that the faster-migrating bands recognized by the polyclonal anti-FUS serum in parental 32Dcl3 cells (Fig. 1C, lane 1) became undetectable after treatment with the proteasome inhibitors (lanes 5 and 7), further suggesting that they represent cleavage products of FUS. Like the endogenous FUS, levels of the ectopically expressed HA-tagged FUS were also regulated in a proteasome-dependent manner and, as expected, most of the HA-tagged FUS was detected in the nucleus (Fig. 1D, top panel). Interestingly, the FUS-associated hnRNP A1 protein was also protected from proteasome-dependent degradation (Fig. 1D, lower panel) and was not downmodulated in BCR-ABL-expressing cells (data not shown).
Proteasome-mediated proteolysis of FUS does not require its ubiquitination.
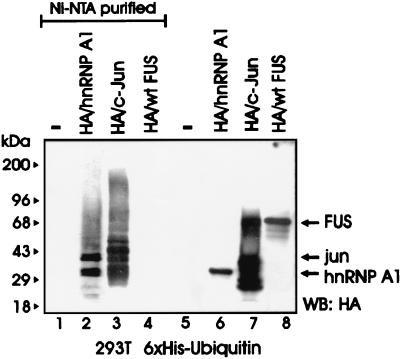
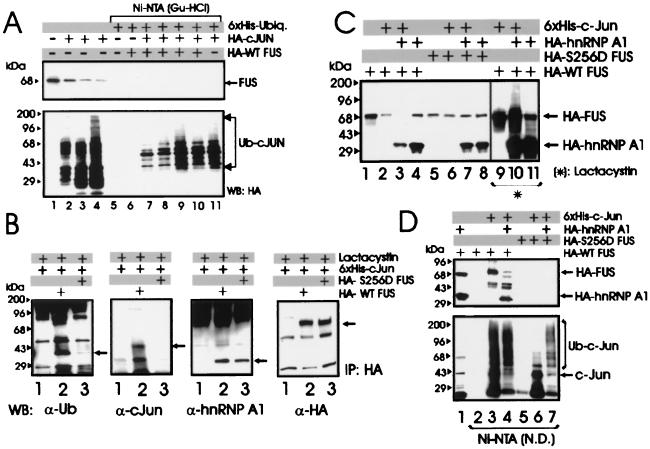
Most cellular proteins targeted for proteasome-dependent degradation undergo an enzymatic modification whereby they are covalently bound to ubiquitin molecules in the form of polyubiquitin chains which function as a degradation signal (31). To determine whether FUS is a substrate for ubiquitination, 293T cells were cotransfected with HA-tagged FUS and His6-tagged ubiquitin plasmids and assessed for ubiquitination. FUS was readily detectable in total lysates (Fig. 2, lane 8) but not in the Ni-NTA-purified fractions (lane 4), suggesting that it was not ubiquitinated at detectable levels. By contrast, the HA-tagged FUS-associated protein hnRNP A1 (42) and the control HA-tagged c-Jun were polyubiquitinated, as indicated by the multiple slowly migrating forms detected by the anti-HA antibody (lanes 2 and 3). FUS polyubiquitination was also undetectable in Western blots using FUS antiserum on anti-HA immunoprecipitates from 32Dcl3 and 32DBCR-ABL cells expressing the HA-tagged ubiquitin (not shown). Accordingly, FUS appears to be one of an unknown number of proteins recognized and degraded by the proteasome without undergoing ubiquitination, although it cannot be excluded that low levels of ubiquitination may contribute to its degradation.
FIG. 2.
FUS proteolysis does not require its ubiquitination. An in vivo ubiquitination assay of c-Jun, hnRNP A1, and FUS was performed. Shown is a Western blot with an anti-HA antibody on nickel chromatography-purified (Ni-NTA resin under denaturing conditions) His6-ubiquitinated proteins (lanes 1 to 4) or on total-cell lysates (lanes 5 to 8) from 293T cells transfected with His6-tagged ubiquitin (lanes 1 to 8) along with HA-hnRNP A1 (lanes 2 and 6), HA-c-Jun (lanes 3 and 7), or HA-FUS (lanes 4 and 8). Data are representative of three independent experiments with similar results.
BCR-ABL-dependent PKCβII activity is required for FUS protein stability.
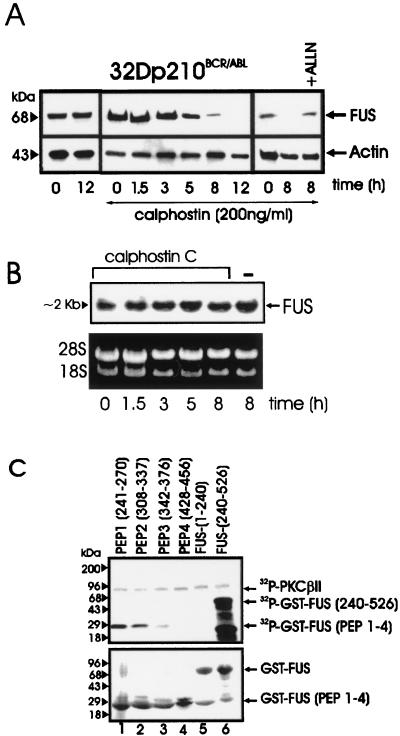
The DNA binding activity of FUS in BCR-ABL-expressing 32Dcl3 cells was inhibited by treatment with a PKCβII-specific inhibitor or upon expression of a PKCβII dominant-negative mutant (30). To determine whether the impairment in FUS binding activity is accompanied by a progressive decline in FUS expression, we assessed FUS levels in BCR-ABL-expressing cells treated with calphostin C, a specific inhibitor of conventional PKCs (39). In IL-3- and serum-starved BCR-ABL-expressing cells treated with a calphostin C concentration which does not induce apoptosis (12), FUS levels were barely detectable at 8 h and undetectable at 12 h, whereas they remained unchanged in untreated cells (Fig. 3A). The decreased FUS expression in calphostin C-treated cells was not due to reduced mRNA levels (Fig. 3B). The proteasome inhibitor ALLN rescued FUS protein expression in 8-h calphostin C-treated BCR-ABL-expressing 32Dcl3 cells (Fig. 3A), suggesting that PKCβII protects FUS from proteasome-mediated degradation. The potential role of PKCβII phosphorylation in FUS stability was further investigated by identifying potential phosphorylation sites and by assessing the properties of proteins carrying a mutated phosphorylation site. PROSITE database analysis of the FUS protein sequence revealed multiple potential PKC phosphorylation sites clustered between amino acids 240 and 526; thus, GST fusion proteins with various FUS peptides were generated and tested as PKCβII substrates. FUS peptides 1 (amino acids 241 to 270) and 2 (amino acids 308 to 337) were heavily phosphorylated, while phosphorylation of peptides 3 (amino acids 342 to 376) and 4 (amino acids 428 to 456) was barely detectable or undetectable (Fig. 3C, lanes 1 to 4). Consistent with the results of a previous study (30), the C terminus but not the N terminus FUS was highly phosphorylated (lanes 5 and 6).
FIG. 3.
PKC-dependent FUS expression and identification of FUS PKCβII phosphorylation sites. (A) Western blot of FUS expression in BCR-ABL-expressing 32Dcl3 cells untreated or treated for the indicated times with calphostin C, alone or in the presence of the proteasome inhibitor ALLN. Actin expression was used as a control. (B) Northern blot of FUS expression in calphostin C-treated (1.5 to 8 h) BCR-ABL-expressing 32Dcl3 cells. (C) In vitro kinase assay (top panel) with recombinant PKCβII as the active kinase and GST-FUS fusion proteins as the substrate. The N-terminal (amino acids 1 to 240) (lane 5) and the C-terminal (amino acids 240 to 526) (lane 6) regions of FUS and four different FUS peptides (lanes 1 to 4) containing the putative PKC phosphorylation sites fused to GST are visible after Coomassie staining of the SDS-PAGE-fractionated kinase reaction products (bottom panel). Data are representative of three different experiments with similar results.
FUS stability and resistance to proteasome degradation depends on its phosphorylation at serine 256.
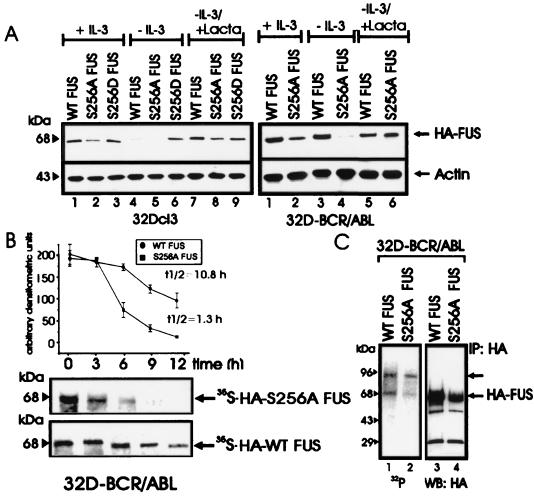
Serine phosphorylation of FUS is required for its DNA binding activity (30), and peptide 1, RGRGGGRGGRGGMGGSDRGGFNKFGGPRDQ (amino acids 241 to 270) which is heavily phosphorylated, contains only one PKCβII phosphorylation site (serine 256 of the SDR motif), while the four PKCβII phosphorylation sites present in peptide 2 do not include serine residues (see Materials and Methods). Accordingly, we generated HA-tagged S256A and S256D FUS mutants to investigate their biochemical properties and susceptibility to proteasome-dependent degradation. Parental 32Dcl3 cells stably transfected with HA-tagged WT FUS or with the S256A mutant showed downmodulated FUS expression after IL-3 starvation, which was restored by treatment with the proteasome inhibitor lactacystin (Fig. 4A, left panel). By contrast, expression of the S256D mutant remained unchanged upon IL-3 removal and after inhibition of proteasome activity (Fig. 4A, left panel). As expected, levels of WT FUS in transfected BCR-ABL-expressing cells were not altered after 8 to 10 h in IL-3- and serum-deprived cultures; under the same culture conditions, expression of the S256A FUS mutant was markedly impaired but was restored by lactacystin (Fig. 4A, right panel). Consistent with the propensity of the S256A FUS mutant to undergo degradation upon serum and IL-3 deprivation of BCR-ABL-expressing cells, the t1/2 of newly synthesized S256A FUS was considerably shorter (t1/2 ≈ 1.5 h) than that of WT FUS (t1/2 ≈ 10.8 h) (Fig. 4B). The S256A mutant retained the ability to associate with PKCβII (data not shown) but was less phosphorylated in vivo (approximately 50%) than was WT FUS (Fig. 4C), suggesting that serine 256 is also an in vivo phosphorylation site. Moreover, the S256A FUS mutant lost the ability to bind DNA (data not shown), and the relative amount of FUS in complex with hnRNP C1/2 and hnRNP A1, two FUS-associated proteins (42), was larger in the anti-HA immunoprecipitates from S256A FUS-expressing cells than from WT FUS- or S256D FUS-expressing cells (data not shown), suggesting that FUS function is regulated by serine 256 phosphorylation.
FIG. 4.
Role of serine 256 in expression, stability, proteasome-mediated degradation, and in vivo phosphorylation of FUS. (A) HA-FUS levels in parental (left panel) and BCR-ABL-expressing (right panel) 32Dcl3 cells stably expressing WT FUS or the S256A or S256D mutant. Cells were maintained in the presence of IL-3 or were IL-3 deprived (for 8 h) in the presence or absence of the proteasome inhibitor lactacystin (Lacta). Actin levels were used as a control. (B) Stability of newly synthesized WT FUS and S256A FUS mutant in IL-3- and serum-deprived (8 h) 32DBCR-ABL cells. Each point of the graph represents the mean and standard deviation of the relative amounts of WT FUS and S256A FUS during the chase period. Values on the graph are representatives of three independent experiments. (C) FUS phosphorylation in in vivo 32P-labeled WT FUS- and S256A-expressing 32DBCR-ABL cells (lanes 1 and 2), and amount of immunoprecipitated (IP) WT and S256A FUS (lanes 3 and 4). Data are representative of three experiments with similar results.
c-Jun requirement for proteasome-dependent FUS proteolysis.
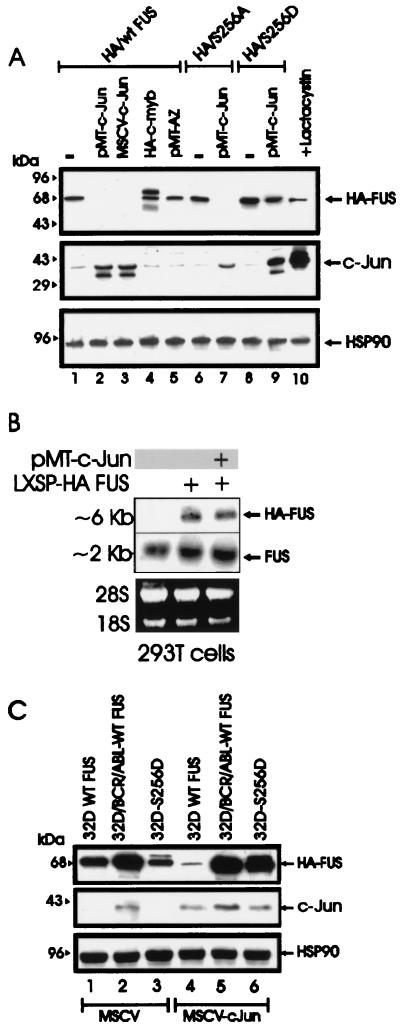
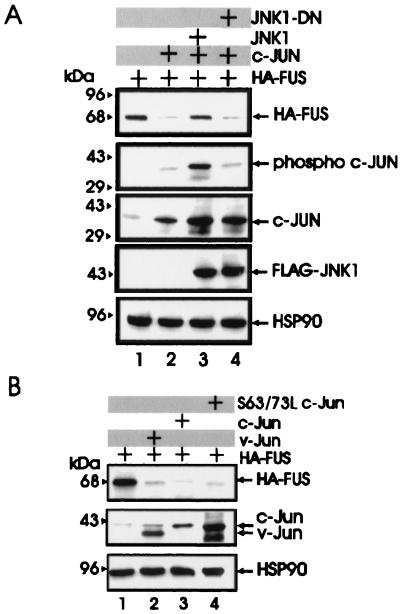
With the exception of the antizyme-dependent degradation of (ODC) (1), the mechanisms involved in the proteasome-dependent degradation of nonubiquitinated substrates are largely undefined. Upon formation of a heterodimeric complex, c-Jun induces the ubiquitination and the proteasome-dependent degradation of the associated ATF2 protein (14). In experiments assessing the potential role of antizyme and c-Jun in FUS degradation, we found that antizyme expression had no effect on FUS levels in transfected 293T cells (Fig. 5A, lane 5) while FUS degradation was induced by expression of two different plasmids carrying the His6-tagged c-Jun cDNA (Fig. (lanes 2 and 3) and it was prevented when the cotransfected cells were treated with the proteasome inhibitor lactacystin (lane 10). Moreover, c-Jun expression promoted the proteolysis of the S256A but not S256D FUS mutant (lanes 6 to 9), suggesting that the effects of c-Jun are specific and that dephosphorylation of serine 256 is a prerequisite for c-Jun-mediated FUS degradation by the proteasome machinery. Overexpression of the HA-tagged c-Myb, used as control for the effect of an ectopic protein on FUS expression, did not alter FUS levels (lane 4). Of note, overexpression of c-Jun had no effect on the mRNA levels of endogenous and exogenous FUS (Fig. 5B). To determine whether BCR-ABL prevents c-Jun-induced FUS degradation, expression of the HA-tagged FUS was assessed in parental and BCR-ABL cells overexpressing WT FUS, as well as in 32Dcl3 cells expressing the S256D FUS mutant, 48 h after infection with a retrovirus carrying the full-length c-Jun cDNA. Exogenous WT FUS expression was markedly downmodulated by c-Jun in parental cells but not in BCR-ABL-expressing 32Dcl3 cells (Fig. 5C, lanes 4 and 5). Moreover, c-Jun overexpression had no effect on the levels of S256D FUS in 32Dcl3 cells expressing this mutant (lane 6). The c-Jun-dependent degradation of WT FUS in 293T cells was blocked by coexpression of c-Jun NH2-terminal kinase 1 (JNK1) but not by a dominant-negative JNK1 (15) (Fig. 6A), suggesting that expression of nonphosphorylated c-Jun is required for proteasome-mediated FUS proteolysis. Overexpression of v-Jun, which is unable to associate with JNKs and cannot be ubiquitinated in vivo (9, 39), induced FUS degradation (Fig. 6B, lane 2), further indicating that this effect is not dependent on Jun-JNK interaction or c-Jun ubiquitination. In addition, expression of S63/73L c-Jun, a mutant which is defective in transactivation (16, 32), induced FUS proteolysis as effectively as did wild-type c-Jun (lanes 3 and 4), suggesting that FUS degradation is not mediated by protein(s) whose expression is transcriptionally regulated by c-Jun.
FIG. 5.
c-Jun requirement for FUS proteasome-dependent degradation. (A) HA-FUS expression (top panel) in transiently transfected 293T cells (lane 1), cotransfected with two different c-Jun expression plasmids (pMT-c-Jun [pMT35] and MSCV-c-Jun) (lanes 2 and 3, respectively), with pMT HA-c-Myb (lane 4), or with a CMV-based vector containing the full-length antizyme (AZ) cDNA (lane 5). Expression of S256A FUS and S256D FUS mutants upon transient transfection in 293T cells (lanes 6 and 8) or cotransfection with pMT-c-Jun (lanes 7 and 9) is also shown. 293T cells were also cotransfected with WT HA FUS and pMT-c-Jun and treated for 8 h with the proteasome inhibitor lactacystin before being subjected to lysis (lane 10). c-Jun (middle panel) and HSP90 (bottom panel) expression were monitored as controls. (B) Ectopic (top panel) and endogenous (middle panel) FUS mRNA expression in parental 293T cells (lane 1) or in cells transfected with the LXSP HA-FUS plasmid alone (lane 2) or cotransfected with pMT-c-Jun (lane 3). Ethidium bromide staining of rRNA is shown as a control for equal loading (bottom panel). (C) Effect of transient expression of c-Jun (middle panel) on HA-FUS levels (top panel) in retrovirus-infected parental or BCR-ABL-expressing 32Dcl3 cells constitutively expressing HA-tagged WT FUS or S256D FUS. HSP90 levels (bottom panel) were monitored as a control of equal loading.
FIG. 6.
Effect of JNK1, v-Jun, and S63/73L c-Jun mutant on FUS expression. (A) HA-FUS expression (top row) in lysates of 293T cells transfected with WT HA-FUS alone (lanes 1 to 4) or with c-Jun (lanes 2 to 4), FLAG-tagged WT JNK1 (lane 3), or a FLAG-tagged dominant-negative JNK1 (lane 4). Phospho-c-Jun levels (second row) were detected using an anti-phospho-Jun antibody (Santa Cruz Biotechnology, Inc.). Total c-Jun levels (third row) were monitored using a mix (1:1) of polyclonal anti-c-Jun antibodies (Santa Cruz Biotechnology and Oncogene Sciences). Levels of exogenous JNK1 (fourth row) were detected using an anti-FLAG antibody (Sigma). (B) HA-FUS expression (top row) in lysates of 293T cells transfected with WT HA-FUS alone (lanes 1 to 4) or with v-Jun (lane 2), c-Jun (lane 3), or the transactivation-deficient S63/73L c-Jun mutant (lane 4). c-Jun and v-Jun levels (second row) were detected using the polyclonal anti-c-Jun antibodies mix described for panel A. HSP90 levels were monitored as control for equal loading.
FUS–hnRNP A1–c-Jun interaction is required for proteasome-dependent FUS proteolysis.
In 293T cells, the ectopically expressed HA-tagged FUS was no longer detectable 48 h after coexpression with c-Jun (Fig. 5), and Western blots with the anti-FUS antibody showed that c-Jun also induced degradation of the endogenous FUS in a time-dependent manner (Fig. 7A, lanes 1 to 4).
FIG. 7.
Role of ubiquitination and of hnRNP A1 expression in c-Jun-induced FUS degradation. (A) Endogenous FUS levels in 293T cells that were not transfected (lane 1) or transfected with HA-c-Jun and harvested 16, 24, and 36 h after transfection (lanes 2 to 4), and an in vivo ubiquitination assay in 293T cells cotransfected with His6-tagged ubiquitin, HA-c-Jun, and HA-WT FUS expression plasmids and harvested 16, 24, 36, and 48 h after transfection (lanes 7 to 10). Western blot with anti-FUS (upper panel) or anti-HA (lower panel) antibody on total-cell lysate (lanes 1 to 4) or Ni-NTA-purified proteins (lanes 5 to 11) from nontransfected cells (lane 1), cells transfected with HA-c-Jun only (lanes 2 to 4), cells transfected with His6-tagged ubiquitin (lanes 5 to 11), plus HA-WT FUS (lane 6), cotransfected with HA-WT FUS and HA-c-Jun (lanes 7–10), or plus HA-c-Jun only (lane 11). (B) Identification of WT and S256D FUS-associated proteins in lactacystin-treated 293T cells. Shown are Western blots with anti-ubiquitin (first panel), anti-c-Jun (second panel), anti-hnRNP A1 (third panel), and anti-HA (fourth panel) antibody on HA-immunoprecipitates (IP) from lysates of 293T cells transfected with His6-tagged c-Jun (lanes 1 to 3) alone (lane 1) or with HA-tagged WT FUS (lane 2) or S256D FUS (lane 3) and treated for 8 h with 10 μM lactacystin before being subjected to lysis. (C) Effect of hnRNP A1 on c-Jun-induced degradation of FUS. Shown are Western blots with an anti-HA antibody on total-cell lysates from 293T cells transfected (1:1:1 molar ratio) with HA-tagged WT FUS (lanes 1 to 4 and 9 to 11) or S256D FUS mutant (lanes 5 to 8), plus His6-tagged c-Jun (lanes 2, 3, 6, 7, 9, and 10) or HA-tagged hnRNP A1 (lanes 3, 4, 7, 8, 10, and 11) left untreated (lanes 2 to 8) or treated (lanes 9 to 11) with the proteasome inhibitor lactacystin. c-Jun and HSP90 levels were measured as internal controls of transfection efficiency and equal loading (data not shown). (D) Ni-NTA pull-down assay performed with the same lysate (1.5 mg) used in the experiment in Fig. 6C. Shown are Western blots with an anti-HA (upper panel) or anti-c-Jun (lower panel) antibody on total-cell lysates (lane 1) or on nondenatured (N.D.) Ni-NTA-purified fractions (lanes 2 to 7) from 293T cells transfected with HA-tagged WT FUS (lanes 1 to 4) or S256D FUS (lanes 5 to 7) along with His6-tagged c-Jun (lanes 3, 4, 6, and 7) or HA-hnRNP A1 (lanes 1, 4, and 7). Data are representative of three experiments with similar results.
If ubiquitination of FUS is important for its degradation, it might be promoted by c-Jun expression and/or its physical interaction with c-Jun, as reported for the transcription factor ATF2 (14). To assess whether c-Jun expression is required for the induction of FUS ubiquitination, 293T cells were transfected with the His6-tagged ubiquitin in the presence of HA-tagged c-Jun and HA-tagged WT FUS in a 1:1 molar ratio. At 16, 24, 36, and 48 h posttransfection, the formation of FUS-ubiquitin conjugates was determined by nickel chromatography performed under denaturing conditions. Indeed, Western blotting with the anti-FUS antibody did not detect ubiquitinated forms of FUS, whereas ubiquitinated c-Jun was readily detectable (Fig. 7A, lanes 5 to 11).
By the Ni-NTA pull-down assay, WT FUS, but not the S256D FUS mutant, was found in complex with an ubiquitinated protein(s) (data not shown), suggesting that this FUS-associated protein might serve as a chaperon for targeting FUS to the 26S proteasome. The nature of this protein was further investigated by Western blotting using an antiubiquitin antibody on HA immunoprecipitates from 293T cells transiently transfected with HA-tagged WT FUS or S256D FUS mutant and His6-tagged c-Jun and treated with 10 μM lactacystin to prevent proteasome-dependent FUS degradation. Indeed, a ubiquitinated protein was readily detected in complex with WT FUS and to a lesser extent with the S256D FUS mutant (Fig. 7B, lanes 2 and 3, α-Ub panel). In the same immunoprecipitates, WT FUS, but not the phosphomimetic S256D FUS mutant, was detected in association with c-Jun (Fig. 7B, lanes 2 and 3, α-cJun panel), consistent with the notion that lack of phosphorylation at serine 256 is required for proteasome targeting and degradation of FUS. Of note, WT FUS or its S256D phosphomimetic mutant were found in complex with the hnRNP A1 protein (Fig. 7B, α-hnRNP A1 panel). Since proteolysis of hnRNP A1 protein is ubiquitin dependent (Fig. 2), it is conceivable that the ubiquitinated protein which interacts with FUS and may be required for FUS degradation represents a ubiquitinated form of hnRNP A1. To address this possibility, 293T cells were transiently transfected with the HA-tagged WT or S256D FUS together with the His6-tagged c-Jun and the HA-tagged hnRNP A1 (1:1:1 molar ratio). Compared to the effect of c-Jun alone (∼75% decrease in FUS levels), coexpression of hnRNP A1 and c-Jun enhanced the degradation of FUS (∼95% decrease in FUS levels) (Fig. 7C, lanes 1 to 4). As expected, overexpression of hnRNP A1 and c-Jun had no effect on the levels of the S256D FUS mutant (lanes 5 to 8). Like FUS, hnRNP A1 was also susceptible to the degradation-promoting effect of c-Jun (lanes 3 and 4), and the effect was even more pronounced at a 1:1:0.5 molar ratio of c-Jun, FUS, and hnRNP A1, respectively (data not shown). In control experiments, c-Jun had no effect on the levels of ectopically expressed c-Myb, which has a short half-life and, like hnRNP A1, undergoes ubiquitin-dependent proteasome degradation (data not shown). c-Jun-dependent FUS and hnRNP A1 degradation was prevented by the proteasome inhibitor lactacystin (10 μM) (Fig. 7C, lanes 9 to 11), which also allowed the detection of poly-ubiquitinated hnRNP A1 protein (lane 10). To exclude the possibility that c-Jun directly targets hnRNP A1 to ubiquitin-dependent proteasome degradation, Ni-NTA pull-down experiments were performed with 1.5 mg of the same lysates of 293T cells expressing the His6-tagged c-Jun and WT FUS or S256D FUS with or without the HA-tagged hnRNP A1 protein (Fig. 7C). c-Jun was found in association with hnRNP A1 upon coexpression with WT FUS (Fig. 7D, lane 4) but not with the S256D FUS mutant (lane 7).
PKCβII-dependent phosphorylation of FUS serine 256 regulates survival and differentiation of myeloid precursor 32Dcl3 cells.
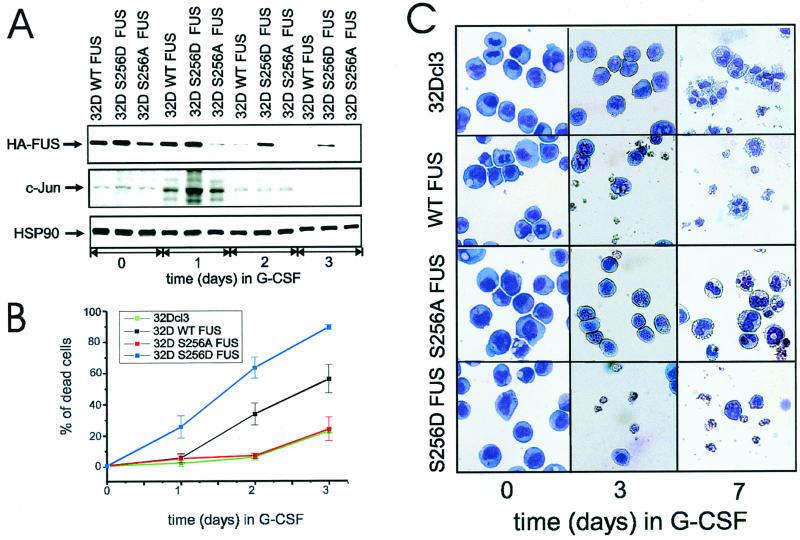
Downregulation of FUS expression accelerates G-CSF-induced granulocytic differentiation of myeloid precursor 32Dcl3 cells, whereas overexpression of FUS induces apoptosis and consequently reduces the number of differentiated cells (30). To assess the role of FUS serine 256 phosphorylation during G-CSF-dependent differentiation, parental 32Dcl3 cells or cells expressing the wild-type (32D WT FUS), the phosphomimetic (32D S256D FUS), or the nonphosphorylatable (32D S256A FUS) FUS were cultured for 7 days in medium containing G-CSF and monitored for FUS expression, survival, and differentiation.
Consistent with the results of a previous study (29), WT HA FUS levels were completely downmodulated after 3 days in G-CSF (Fig. 8A, top panel); by contrast, S256A FUS expression was suppressed within 24 h (Fig. 8A, top panel), while the levels of the phosphomimetic FUS (S256D) were almost unaffected after 2 days and remained readily detectable after 3 days of treatment with G-CSF (Fig. 8A, top panel). Of interest, downregulation of FUS levels temporally correlated with G-CSF-induced c-Jun expression, which peaked at 24 h and became undetectable after 3 days of exposure to G-CSF (Fig. 8A, middle panel), consistent with the involvement of c-Jun in the induction of FUS degradation. Interestingly, overexpression of the S256D FUS mutant was more potent than that of WT FUS in inducing apoptosis of 32Dcl3 cells growing in G-CSF-containing medium (Fig. 8B and C). Moreover, WT FUS-expressing cells escaping apoptosis were able to undergo terminal differentiation, while surviving S256D FUS-expressing cells remained undifferentiated (Fig. 8C) and proliferated in G-CSF-containing medium (data not shown). Conversely, mutation of FUS serine 256 to alanine abolished the apoptotic effects of FUS, allowing an apparently normal granulocytic differentiation (Fig. 8B and C). Together, these data support a model in which FUS degradation, possibly enhanced by c-Jun, is required for granulocytic differentiation of 32Dcl3 cells.
FIG. 8.
Effect of Ser 256 FUS mutant expression on G-CSF-induced differentiation of 32Dcl3 cells. (A) Kinetics of FUS, c-Jun, and HSP90 expression (Western blotting) in a representative (of three for each transfectant) clone of WT FUS, S256D FUS, or S256A FUS-expressing 32Dcl3 cells cultured in the presence of G-CSF for 0, 1, 2, or 3 days. (B) Effect of G-CSF on the viability of parental and derivative cell lines ectopically expressing WT FUS, S256A FUS, or S256D FUS. Each point represents the average of three independent experiments and standard deviation. The percentage cell death was determined by trypan blue exclusion. (C) G-CSF-induced differentiation of parental and representative (of three for each transfectant) 32Dcl3-derived cell lines. Representative micrographs of May-Grunwald-Giemsa-stained cytospins are shown.
DISCUSSION
The observation that BCR-ABL induces FUS expression and binding to nucleic acids (30) led us to study the mechanisms controlling FUS turnover in normal and BCR-ABL-expressing hematopoietic cells. Overexpression of FUS in BCR-ABL-expressing cells reflects both increased mRNA levels and enhanced protein stability (Fig. 1). However, treatment of BCR-ABL-expressing cells with the protein synthesis inhibitor cycloheximide did not alter FUS DNA binding activity or its expression (data not shown), suggesting that the primary mechanism for the increase in FUS expression is posttranslational and unlikely to depend on BCR-ABL-regulated pathways leading to enhanced FUS transcription. The degradation of FUS in IL-3-deprived myeloid 32Dcl3 cells was specifically rescued by proteasome inhibitors but was not dependent on its polyubiquitination. Although technical limitations may prevent the detection of low levels of ubiquitination, it is conceivable that FUS, like other proteins whose prototype is ODC (1, 27), undergoes proteasome-dependent degradation without prior ubiquitination. Probably, the association with adapter molecules, like the antizyme for ODC, targets these proteins for degradation (31). However, the cofactors that might promote the proteasome-dependent degradation of two of these proteins, the nuclear factors c-Jun (unconjugated form) (17) and SP1 (38), are still unknown.
The association of various kinases with their substrates is required in several cases of ubiquitin-proteasome degradation, and the mechanism responsible for triggering this process is, in most cases, dependent on recognition of the phosphorylated substrate (13). However, phosphorylation may also prevent the degradation of some substrates, such as c-Jun, ATF2, and p53 (13, 27). FUS phosphorylation by PKCβII appears to prevent its proteasome-dependent proteolysis. Indeed, the S256D FUS mutant in which the serine phosphorylation site was replaced with the phosphomimetic aspartic acid was less susceptible to degradation induced by IL-3-deprivation, while the S256A mutant was even more susceptible to degradation than was WT FUS (Fig. 4). Expression of the S256A mutant in BCR-ABL-expressing cells was lower than that of WT FUS (several S256A-expressing clones were analyzed), downmodulated upon IL-3 and serum starvation, and restored upon treatment with the proteasome inhibitor lactacystin (Fig. 4). Although PKCβII was still able to associate with S256A FUS and this mutant partially retained the ability to be phosphorylated in vivo (Fig. 4), the serine-to-alanine mutation at amino acid 256 markedly altered the half-life of FUS (Fig. 4), influenced its ability to associate with hnRNP C1/2 and A1, suppressed its DNA binding activity, and had no effect on the ability of G-CSF to induce the differentiation of 32Dcl3 cells (Fig. 8C). The location of the Ser 256 PKCβII phosphorylation site within the first RGG box, reportedly involved in both RNA-protein and protein-protein interaction (2, 5), raises the possibility that an interacting protein associates with FUS when Ser 256 is not phosphorylated and promotes its proteasome-dependent degradation. Unlike ODC, FUS degradation did not depend on expression of the ODC-cofactor antizyme but was induced by c-Jun, previously shown to promote the ubiquitin- and proteasome-dependent degradation of the transcription factor ATF2 (14). The c-Jun-dependent degradation of FUS was suppressed by expression of Jun kinase 1 but not of a Jun kinase dominant negative mutant, suggesting that a nonphosphorylated c-Jun is required for the effect; as expected, a transactivation-defective c-Jun mutant (S63/73L) (32) was highly effective in inducing FUS degradation, suggesting that c-Jun-dependent gene expression is not required for the effect. Moreover, since v-Jun lacks the c-Jun δ domain responsible for the interaction with JNK/SAPK and for c-Jun ubiquitination (9, 40), its ability to induce FUS proteolysis suggests that such an effect is not dependent on c-Jun association with Jun kinase and/or on c-Jun ubiquitination.
c-Jun targeting of FUS for proteasome-mediated degradation was dependent on the phosphorylation status of FUS Ser 256, since the S256D FUS mutant was resistant to c-Jun-induced proteolysis (Fig. 5). As expected, WT-FUS levels in BCR-ABL-expressing cells were not altered by c-Jun overexpression, confirming the role of BCR-ABL as a regulator of FUS stability via the induction of PKCβII-dependent phosphorylation and, probably, via its ability to activate Jun kinase (34). Unlike the c-Jun-dependent degradation of ATF2, which requires ATF2 ubiquitination induced by formation of the c-Jun–ATF2 complex (14), measurable levels of ubiquitinated FUS were not detected (Fig. 7). Although not ubiquitinated at detectable levels, WT FUS and, to a lesser extent, the phosphomimetic FUS mutant were found in association with a ubiquitinated protein of ∼40 kDa (Fig. 7), suggesting that this FUS-associated protein might be responsible for directing FUS to the proteasome. Since WT FUS interacts with nonubiquitinated c-Jun (Fig. 7) and since the nonubiquitinable v-Jun (40) induces FUS degradation (Fig. 6), it seems unlikely that c-Jun itself targets FUS to the proteasome. c-Jun may, however, favor the posttranslational modification(s) (i.e., ubiquitination) of this FUS-associated protein, providing a signal for targeting FUS to the proteasome. hnRNP A1, a FUS-associated protein (35), is polyubiquitinated and undergoes proteasome-dependent degradation in a c-Jun-dependent manner. These findings, along with the observation that hnRNP A1 enhances the c-Jun-induced degradation of FUS (Fig. 7), suggest that the association with ubiquitinated hnRNP A1 represents one of the mechanisms for targeting FUS to the proteasome. In fact, WT FUS was found in complex with c-Jun and hnRNP A1, whereas the S256D FUS mutant, although able to associate with hnRNP A1, did not interact with c-Jun (Fig. 7). Since c-Jun itself does not interact with hnRNP A1 (Fig. 7), it seems likely that, directly or indirectly, it associates with the FUS-hnRNP A1-containing complex promoting the proteasome-dependent degradation of both FUS and hnRNP A1. Levels of c-Jun were higher in cells cotransfected with degradation-resistant S256D FUS than with degradation-prone WT FUS (data not shown), suggesting that c-Jun in complex with FUS and hnRNPA1 also undergoes degradation.
Although it has been reported that the proteasome specifically degrades only the ubiquitinated subunits of a multiprotein complex (19), there is also evidence that monoubiquitinated proteins (20) and nonubiquitinated proteins or protein complexes, like ODC-antizyme, are efficiently degraded by the 26S proteasome (31). Thus, the formation of a complex with a ubiquitinated protein(s) might be sufficient for the proteasome degradation of nonubiquitinated FUS.
Since expression of hnRNP A1 is abundant whereas c-Jun levels are modulated during the cell cycle or upon induction of differentiation, hnRNP A1 might have primarily a “chaperone” function for FUS degradation whereas the c-Jun-dependent effects on FUS stability might be functionally significant during these processes. Consistent with previous studies (21), c-Jun expression is induced by G-CSF treatment of 32Dcl3 cells (Fig. 8) and precedes the downmodulation of FUS, an event required for granulocytic differentiation (30). Interestingly, the ectopically expressed proteolysis-resistant S256D FUS mutant was only partly downmodulated during G-CSF-induced differentiation of 32Dcl3 cells and caused massive apoptosis and the emergence of a cohort of differentiation-arrested cells growing in G-CSF-containing medium. In the apoptosis-resistant BCR-ABL-expressing 32Dcl3 cells, failure to downmodulate FUS levels might be one of the mechanisms preventing G-CSF-induced differentiation, thereby contributing to leukemogenesis.
In addition to PKCβII-dependent phosphorylation of FUS at Ser 256, oncogenic BCR-ABL might suppress FUS proteolysis by causing an increase in c-Jun phosphorylation mediated by active Jun kinase 1. Since activation of the Jun kinase pathway is required for BCR-ABL-dependent transformation (11, 34), it is possible that the oncogenic effects of active Jun kinase are mediated by stabilization of FUS and, perhaps, of other nuclear regulators.
In summary, FUS expression is, in part, controlled by a process of proteasome-mediated degradation regulated by PKCβII-dependent phosphorylation, c-Jun expression, and possibly hnRNP A1 ubiquitination. The execution of these processes appears to be important for granulocytic differentiation, while its suppression by oncogenic BCR-ABL might contribute to BCR-ABL-dependent leukemogenesis. This might be especially relevant in chronic myelogenous leukemia blast crisis, in which BCR-ABL-expressing cells are differentiation arrested and exhibit abundant FUS expression (30).
ACKNOWLEDGMENTS
Angela Iervolino and Vincenzo Cesi contributed equally to this work.
We thank R. Trotta, P. Salomoni, M. Prisco, F. Peruzzi, and F. Condorelli for helpful discussions. We thank Yan Fu for assistance in production of the anti-FUS polyclonal antibody.
D. Perrotti was supported in part by a fellowship from the American-Italian Foundation for Cancer Research (New York, N.Y.). This work was supported in part by NIH grants to B. Calabretta.
REFERENCES
- 1.Bercovich Z, Rosenberg-Hasson Y, Ciechanover A, Kahana C. Degradation of ornithine decarboxylase in reticulocyte lysate is ATP-dependent but ubiquitin-independent. J Biol Chem. 1989;264:15949–15952. [PubMed] [Google Scholar]
- 2.Bouvet P, Diaz J J, Kindbeiter K, Madjar J J, Amalric F. Nucleolin interacts with several ribosomal proteins through its RGG domain. J Biol Chem. 1998;273:19025–19029. doi: 10.1074/jbc.273.30.19025. [DOI] [PubMed] [Google Scholar]
- 3.Bureau J P, Henry L, Baz A, Scherrer K, Chateau M T. Prosome (proteasome) changes during differentiation are related to the type of inducer. Mol Biol Rep. 1997;24:57–62. doi: 10.1023/a:1006856707793. [DOI] [PubMed] [Google Scholar]
- 4.Campanero M R, Flemington E K. Regulation of E2F through ubiquitin-proteasome-dependent degradation: stabilization by the pRB tumor suppressor protein. Proc Natl Acad Sci USA. 1997;94:2221–2226. doi: 10.1073/pnas.94.6.2221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cartegni L, Maconi M, Morandi E, Cobianchi F, Riva S, Biamonti G. hnRNP A1 selectively interacts through its Gly-rich domain with different RNA-binding proteins. J Mol Biol. 1996;259:337–348. doi: 10.1006/jmbi.1996.0324. [DOI] [PubMed] [Google Scholar]
- 6.Ciechanover A. The ubiquitin-proteasome pathway: on protein death and cell life. EMBO J. 1998;17:7151–7160. doi: 10.1093/emboj/17.24.7151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cortez D, Kadlec L, Pendergast A M. Structural and signaling requirements for BCR/ABL-mediated transformation and inhibition of apoptosis. Mol Cell Biol. 1995;15:5531–5541. doi: 10.1128/mcb.15.10.5531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Crozat A, Aman P, Mandahl N, Ron D. Fusion of CHOP to a novel RNA-binding protein in human myxoid liposarcoma. Nature. 1993;363:640–644. doi: 10.1038/363640a0. [DOI] [PubMed] [Google Scholar]
- 9.Dai T, Rubie E, Franklin C C, Kraft A, Gillespie D A, Avruch J, Kyriakis J M, Woodgett J R. Stress-activated protein kinases bind directly to the delta domain of c-Jun in resting cells: implications for repression of c-Jun function. Oncogene. 1995;10:849–855. [PubMed] [Google Scholar]
- 10.Dai Z, Quackenbush R C, Courtney K D, Grove M, Cortez D, Reuther G W, Pendergast A M. Oncogenic Abl and Src tyrosine kinases elicit the ubiquitin-dependent degradation of target proteins through a Ras-independent pathway. Genes Dev. 1998;12:1415–1424. doi: 10.1101/gad.12.10.1415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dickens M, Rogers J S, Cavanagh J, Raitano A B, Xia Z, Halpern J R, Greenberg M E, Sawyers C L, Davis R J. A cytoplasmic inhibitor of the JNK signal transduction pathway. Science. 1997;277:690–693. doi: 10.1126/science.277.5326.693. [DOI] [PubMed] [Google Scholar]
- 12.Evans C A, Lord J M, Owen-Lynch P J, Johnson G, Dive C, Whetton A D. Suppression of apoptosis by v-ABL protein tyrosine kinase is associated with nuclear translocation and activation of protein kinase C in an interleukin-3-dependent haemopoietic cell line. J Cell Sci. 1995;108:2591–2598. doi: 10.1242/jcs.108.7.2591. [DOI] [PubMed] [Google Scholar]
- 13.Fuchs S Y, Fried V A, Ronai Z. Stress-activated kinases regulate protein stability. Oncogene. 1998;17:1483–1490. doi: 10.1038/sj.onc.1202184. [DOI] [PubMed] [Google Scholar]
- 14.Fuchs S Y, Ronai Z. Ubiquitination and degradation of ATF2 are dimerization dependent. Mol Cell Biol. 1999;19:3289–3298. doi: 10.1128/mcb.19.5.3289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gupta S, Campbell D, Derijard B, Davis R J. Transcription factor ATF2 regulation by the JNK signal transduction pathway. Science. 1995;267:389–393. doi: 10.1126/science.7824938. [DOI] [PubMed] [Google Scholar]
- 16.Hunter T, Karin M. The regulation of transcription by phosphorylation. Cell. 1992;70:375–387. doi: 10.1016/0092-8674(92)90162-6. [DOI] [PubMed] [Google Scholar]
- 17.Jariel-Encontre I, Pariat M, Martin F, Carillo S, Salvat C, Piechaczyk M. Ubiquitinylation is not an absolute requirement for degradation of c-Jun protein by the 26 S proteasome. J Biol Chem. 1995;270:11623–11627. doi: 10.1074/jbc.270.19.11623. [DOI] [PubMed] [Google Scholar]
- 18.Jariel-Encontre I, Salvat C, Steff A M, Pariat M, Acquaviva C, Furstoss O, Piechaczyk M. Complex mechanisms for c-fos and c-jun degradation. Mol Biol Rep. 1997;24:51–56. doi: 10.1023/a:1006804723722. [DOI] [PubMed] [Google Scholar]
- 19.Johnson E S, Gonda D K, Varshavsky A. cis-trans recognition and subunit-specific degradation of short-lived proteins. Nature. 1990;346:287–291. doi: 10.1038/346287a0. [DOI] [PubMed] [Google Scholar]
- 20.Johnson E S, Bartel B, Seufert W, Varshavsky A. Ubiquitin as a degradation signal. EMBO J. 1992;11:497–505. doi: 10.1002/j.1460-2075.1992.tb05080.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kreider B L, Rovera G. The immediate early gene response to a differentiative stimulus is disrupted by the v-abl and v-ras oncogenes. Oncogene. 1992;7:135–140. [PubMed] [Google Scholar]
- 22.Loidl G, Groll M, Musiol H J, Huber R, Moroder L. Bivalency as a principle for proteasome inhibition. Proc Natl Acad Sci USA. 1999;96:5418–5422. doi: 10.1073/pnas.96.10.5418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lugo T G, Pendergast A M, Muller A J, Witte O N. Tyrosine kinase activity and transformation potency of bcr-abl oncogene products. Science. 1990;247:1079–1082. doi: 10.1126/science.2408149. [DOI] [PubMed] [Google Scholar]
- 24.Luscher B, Eisenman R N. c-myc and c-myb protein degradation: effect of metabolic inhibitors and heat shock. Mol Cell Biol. 1988;8:2504–2512. doi: 10.1128/mcb.8.6.2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Matsufuji S, Miyazaki Y, Kanamoto R, Kameji T, Murakami Y, Baby T G, Fujita K, Ohno T, Hayashi S. Analyses of ornithine decarboxylase antizyme mRNA with a cDNA cloned from rat liver. J Biochem (Tokyo) 1990;108:365–371. doi: 10.1093/oxfordjournals.jbchem.a123207. [DOI] [PubMed] [Google Scholar]
- 26.Murakami Y, Matsufuji S, Kameji T, Hayashi S, Igarashi K, Tamura T, Tanaka K, Ichihara A. Ornithine decarboxylase is degraded by the 26S proteasome without ubiquitination. Nature. 1992;360:597–599. doi: 10.1038/360597a0. [DOI] [PubMed] [Google Scholar]
- 27.Musti A M, Treier M, Bohmann D. Reduced ubiquitin-dependent degradation of c-Jun after phosphorylation by MAP kinases. Science. 1997;275:400–402. doi: 10.1126/science.275.5298.400. [DOI] [PubMed] [Google Scholar]
- 28.Panagopoulos I, Mandahl N, Mitelman F, Aman P. Two distinct FUS breakpoint clusters in myxoid liposarcoma and acute myeloid leukemia with the translocations t(12;16) and t(16;21) Oncogene. 1995;11:1133–1137. [PubMed] [Google Scholar]
- 29.Pear W S, Nolan G P, Scott M L, Baltimore D. Production of high-titer helper free retroviruses by transient transfection. Proc Natl Acad Sci USA. 1993;90:8392–8396. doi: 10.1073/pnas.90.18.8392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Perrotti D, Bonatti S, Trotta R, Martinez R, Skorski T, Salomoni P, Grassilli E, Iozzo R V, Cooper D R, Calabretta B. TLS/FUS, a pro-oncogene involved in multiple chromosomal translocations, is a novel regulator of BCR/ABL-mediated leukemogenesis. EMBO J. 1998;17:4442–4455. doi: 10.1093/emboj/17.15.4442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pickart C M. Targeting of substrates to the 26S proteasome. FASEB J. 1997;11:1055–1066. doi: 10.1096/fasebj.11.13.9367341. [DOI] [PubMed] [Google Scholar]
- 32.Pulverer B J, Kyriakis J M, Avruch J, Nikolakaki E, Woodgett J R. Phosphorylation of c-jun mediated by MAP kinases. Nature. 1991;353:670–674. doi: 10.1038/353670a0. [DOI] [PubMed] [Google Scholar]
- 33.Rabbitts T H, Forster A, Larson R, Nathan P. Fusion of the dominant negative transcription regulator CHOP with a novel gene FUS by translocation t(12;16) in malignant liposarcoma. Nat Genet. 1993;4:175–180. doi: 10.1038/ng0693-175. [DOI] [PubMed] [Google Scholar]
- 34.Raitano A B, Halpern J R, Hambuch T M, Sawyers C L. The BCR/ABL leukemia oncogene activates Jun Kinase and requires Jun for transformation. Proc Natl Acad Sci USA. 1995;92:11746–11750. doi: 10.1073/pnas.92.25.11746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ron D. TLS-CHOP and the role of RNA-binding proteins in oncogenic transformation. Curr Top Microbiol Immunol. 1997;220:131–142. doi: 10.1007/978-3-642-60479-9_8. [DOI] [PubMed] [Google Scholar]
- 36.Sawyers C L, Callahan W, Witte O N. Dominant negative MYC blocks transformation by ABL oncogenes. Cell. 1992;70:901–910. doi: 10.1016/0092-8674(92)90241-4. [DOI] [PubMed] [Google Scholar]
- 37.Shimizu K, Ichikawa H, Tojo A, Kaneko Y, Maseki N, Hayashi Y, Ohira M, Asano S, Ohki M. An ets-related gene, ERG, is rearranged in human myeloid leukemia with t(16;21) chromosomal translocation. Proc Natl Acad Sci USA. 1993;90:10280–10284. doi: 10.1073/pnas.90.21.10280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Su K, Roos M D, Yang X, Han I, Paterson A J, Kudlow J E. An N-terminal region of Sp1 targets its proteasome-dependent degradation in vitro. J Biol Chem. 1999;274:15194–15202. doi: 10.1074/jbc.274.21.15194. [DOI] [PubMed] [Google Scholar]
- 39.Tamaoki T. Use and specificity of staurosporine, UCN-01, and calphostin C as protein kinase inhibitors. Methods Enzymol. 1991;201:340–347. doi: 10.1016/0076-6879(91)01030-6. [DOI] [PubMed] [Google Scholar]
- 40.Treier M, Staszewski L M, Bohmann D. Ubiquitin-dependent c-Jun degradation in vivo is mediated by the delta domain. Cell. 1994;78:787–798. doi: 10.1016/s0092-8674(94)90502-9. [DOI] [PubMed] [Google Scholar]
- 41.Wong K K, Zou X, Merrell K T, Patel A J, Marcu K B, Chellappan S, Calame K. v-Abl activates c-myc transcription through the E2F site. Mol Cell Biol. 1995;15:6535–6544. doi: 10.1128/mcb.15.12.6535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zinszner H, Albalat R, Ron D. A novel effector domain from the RNA-binding protein TLS or EWS is required for oncogenic transformation by CHOP. Genes Dev. 1994;8:2513–2526. doi: 10.1101/gad.8.21.2513. [DOI] [PubMed] [Google Scholar]
- 43.Zou X, Calame K. Signaling pathways activated by oncogenic forms of Abl tyrosine kinase. J Biol Chem. 1999;274:18141–18144. doi: 10.1074/jbc.274.26.18141. [DOI] [PubMed] [Google Scholar]