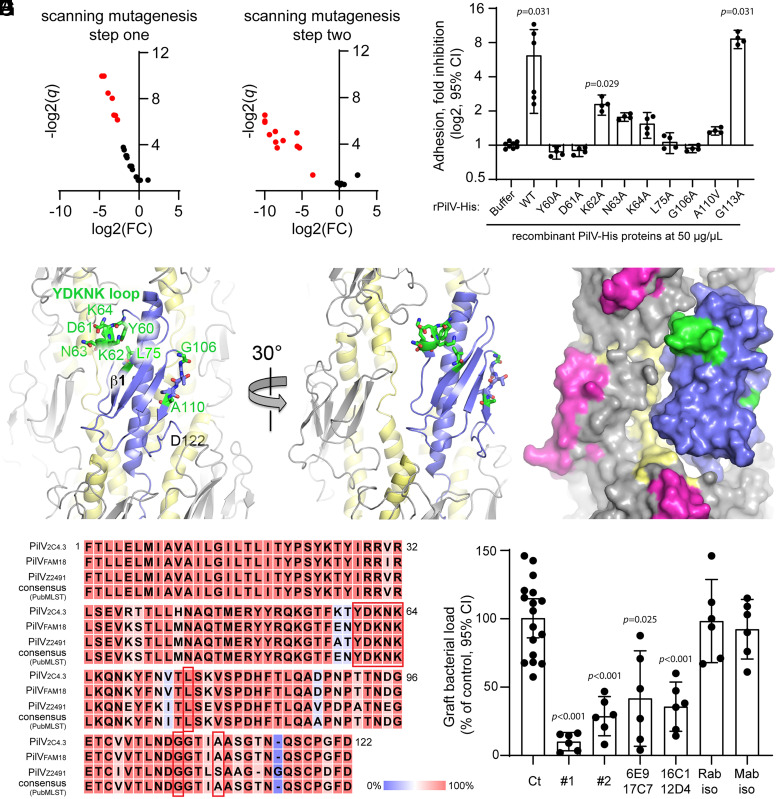
Fig. 4.
PilV residues involved in host cell adhesion. (A and B) Identification of PilV mutant defective for adhesion. Volcano plots, adhesion of N. meningitidis ΔpilV strain expressing PilV variants. Data are shown for mutants in which pili are produced at approximately WT levels and PilV is detected in the pilus preparation. Adhesion is expressed as log2 of fold change [log2(FC)] between mutated and WT PilV, and statistical significance is expressed as −log2(q value) (−log2(q)). Red dots represent mutants defective for adhesion. (A) Mutants selected during the first step of mutagenesis with triplet Ala/Val substitutions; (B) mutants selected during the second step of mutagenesis looking at single Ala/Val substitutions at sites of adhesion defects (SI Appendix, Supplemental Methods). (C) The ability of WT rPilV and rPilV variants to competitively inhibit N. meningitidis adhesion to hCMEC/D3 cells was assessed. Endothelial cells were treated with 50 μg/mL recombinant proteins for 30 min prior to infection with N. meningitidis WT strain for 30 min. The number of adherent cell-associated bacteria was determined by counting CFUs. Data shown are averaged values for two independent experiments. Adhesion is expressed as log2(mean) ± 95% CI of fold inhibition, normalized on bacterial adhesion of cells treated with buffer only. Statistical analyses were performed against buffer using the Kruskal–Wallis test. (D–F) N. meningitidis T4P structure with one PilE subunit replaces with PilV as for Fig. 3. (D) Cartoon representation of the pilus with PilV shown in blue, PilE in gray with α1 colored yellow. Residues implicated in host cell binding are shown in stick representation with carbon atoms in green, nitrogens in blue, and oxygens in red. The YDKNK loop is the Y60-K64 loop. (E and F) Cartoon and space-filling representations of the pilus model rotated about its axis to show how the Y60-K64 loop contacts the neighboring PilE and may form part of a larger receptor binding site. The hypervariable loop, which is part of the protruding β-hairpin of PilE, is colored magenta in F. (G) Alignment of PilV protein sequences from N. meningitidis 2C4.3, FAM18, and Z2491 strains and that of the PubMLST consensus sequence (SI Appendix, Fig. S4). Conserved residues are highlighted in red, and nonconserved residues are highlighted in blue. Residues implicated in host-cell adhesion are boxed in red. (H) Graft bacterial loads of human skin–grafted SCID mice injected intravenously with buffer (control), 100 μg anti-PilV rabbit polyclonal Immunoglobulin G (IgG) anti bodies (#1 and #2), 100 μg anti-PilV peptide mouse monoclonal IgG antibodies (raised against the P80DHFTLQADPNPTTNDGE97 peptide: 6E9 + 17C7 and 16C1 + 12D4) or their respective isotype control antibodies, followed by infection with 5 × 105 CFUs of WT meningococci. Graft bacterial loads were determined 4 h after infection and normalized to that of the control (buffer only). Values shown are the averages of two independent experiments performed with a skin patch from different donors. Each dot represents a single mouse; data are expressed as the mean ± 95% CI of CFU/g. Statistical analyses were performed against the control with the Brown–Forsythe test and Welch’s ANOVA.