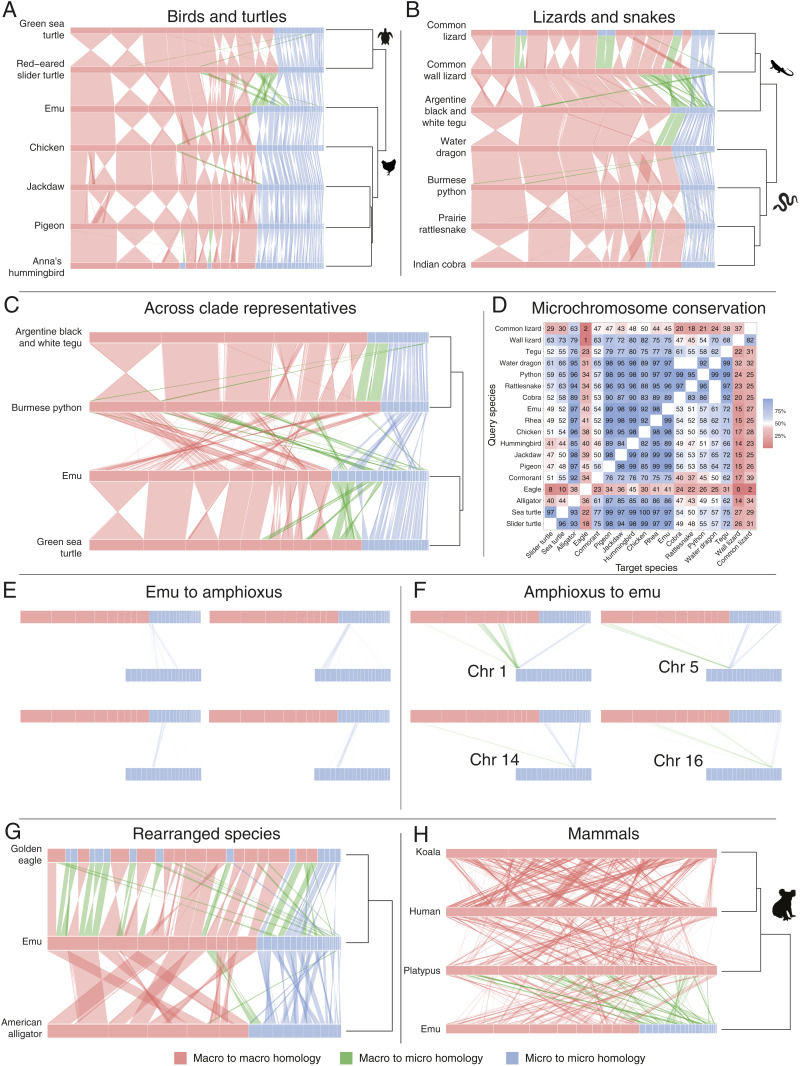
Fig. 3.
Sequence homology plots within and between birds, reptiles, and mammals and comparison to the chordate amphioxus. Relationships between species and genome sizes are shown on the right. Genomes are resized, with chromosome sizes depicted as a proportion of genome length. Some chromosomes have been reordered in the plots to better show homologies within and between clades (see SI Appendix, Table S2). Macrochromosomes are shown in red and microchromosomes in blue; changes between these states are shown in green. Chained and netted alignments were filtered for a minimum length of 100 kb for vertebrate species and 5 kb for amphioxus. Sequence homology between macro- and microchromosomes within (A) birds and turtles and (B) squamates. (C) Sequence homology between macro and microchromosomes of a representative lizard (tegu), snake (python), bird (emu), and the green sea turtle. (D) Heat map showing the fraction (as a function of alignment chain length) of microchromosomes in the query species (y axis) that align to microchromosomes in the target species (x axis). (E and F) Comparisons of emu and amphioxus chromosomes: Single emu microchromosomes have homology to one (or two) amphioxus chromosomes (E), single amphioxus chromosomes detect strong homology to one (or more) emu microchromosomes (from left to right DNAzoo scaffolds 9, 10, 29 and 8), as well as to macrochromosome regions (F). (G) Sequence comparisons between emu and the rearranged alligator and eagle genomes. (H) Sequence comparisons between emu and mammals: eutherian (human), marsupial (koala), and monotreme (platypus)