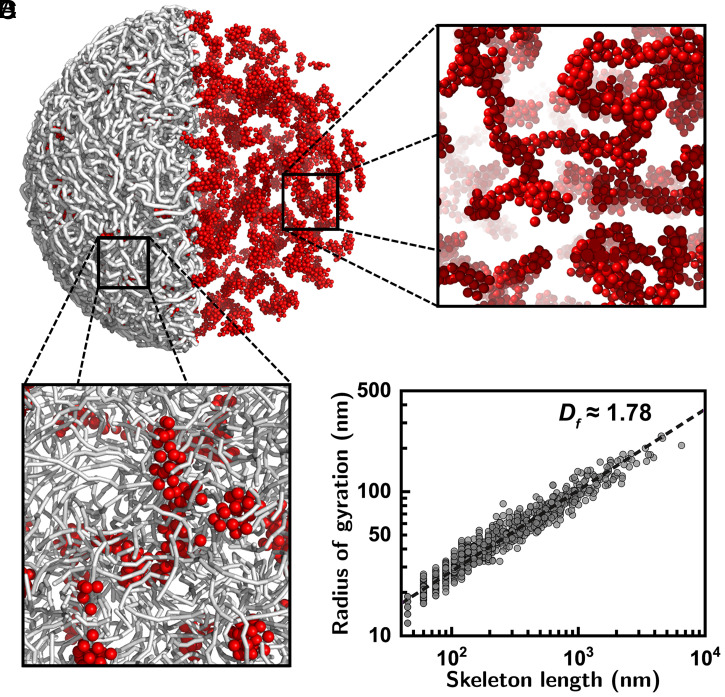
Fig. 4.
Langevin molecular dynamics simulations displaying formation of high-density structures. (A) Molecular dynamics simulation of a coarse-grained polymer model for describing the chromatins and DNA-binding protein complex systems. Here, the gray sticks visualize the chromatin fiber and the red spheres depict the DNA-binding proteins. For further information on the model and simulation details see SI Appendix. Simulations confirmed the formation of high-density structures via aggregations of the proteins and chromatins. For visual inspection of the protein aggregations the chromatin was not displayed in the right hemisphere. (B and C) Zoomed-in views of the snapshot images with (B) and without (C) the chromatin. Molecular dynamics simulations ascertained that the chromatin–protein complex forms a DLCA-like high-density structure. (D) The radii of gyration were estimated at different length scales for the skeletons of the high-density structures from 10 independent simulation runs. The best fit shown in the dashed line leads to , consistent with the experimental results. The obtained is a time-independent stationary value (see vs. simulation time in SI Appendix, section S1.2.