Fig. 5.
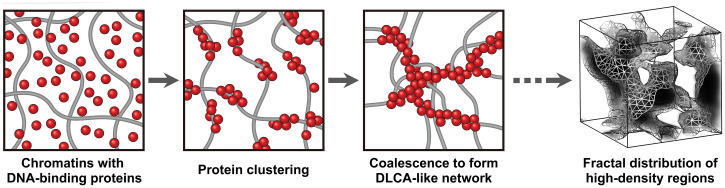
Stochastic chromatin packing with DNA-binding proteins via diffusion-limited cluster aggregation. A mechanistic model describing the chromatin packing process to form randomly distributed high-density structures in the metaphase chromosome is proposed. The complex networks with dispersed chromatins and DNA-binding proteins start to form small clustering with self-aggregation of proteins. With further development of the self-aggregating reaction, enlarged protein clusters becomes coalesced while driving random concentration of chromatins accordingly, which leads to the formation of large-scale chromatin–protein networks following a DLCA structure. As verified by the Langevin molecular dynamics simulations, this stochastically driven chromatin condensation model can describe the fractal distribution of high-density structures observed in metaphase chromosomes.