Abstract
Economic losses due to influenza A virus (IAV) infections are substantial and a global problem, ranking among the top three major health challenges in the swine industry. Currently, H1 and H3 subtypes circulate in pigs globally associated with different combinations of N1 and N2 subtypes; however, the origin, gene constellation, and antigenic makeup of IAV vary greatly on different continents. Vaccination is one means of mitigating the effects of IAV disease, and vaccines are most effective if the strains included closely match the currently circulating strains in pigs. Genetic analyses provide panoramic views of the virus landscape at the sequence level and, thus, can aid in the selection of well-matched swine IAV vaccine strains, but is not sufficient alone. Additionally, a major challenge in selecting appropriate swine IAV vaccine strains is the co-circulation of multiple lineages of viruses in the same region, requiring multivalent or broadly cross-reacting antigens. Due to this complex IAV ecology in swine, new vaccination strategies and vaccine platforms are needed. The hemagglutinin (HA) viral protein is the major target of neutralizing antibodies, which are widely considered to be correlated with protection. Virus variants that are not recognized by previously elicited antibodies can render traditional vaccines that primarily elicit humoral responses ineffective, and therefore result in the need for vaccine strain reformulation and re-vaccination. In the future, new vaccine platforms may be on the market that will provide alternative options to those currently available. Nonetheless, a collaborative approach is needed to improve IAV vaccine strain selection for use in swine.
Keywords: Influenza A virus, Swine, Vaccines, H1N1, H1N2, H3N2
1. Introduction
Economic losses due to swine influenza infections are substantial and a global problem, ranking among the top three major health challenges in the swine industry. Swine influenza is an acute respiratory disease caused by influenza type A virus (IAV) of the family Orthomyxoviridae. The genome of IAV consists of eight RNA strands in the negative sense orientation encoding for at least 12–13 proteins (PB2, PB1, PB1-F2, PB1-N40, PA, PA-X, HA, NP, NA, M1, M2, NS1 and NS2/NEP). Swine influenza infections are often characterized by rapid onset of high fever, lethargy, loss of appetite, labored abdominal breathing, and coughing (Van Reeth et al., 2012). The disease lasts for 2–6 days and although mortality is low and most animals recover, weight loss can be severe. Swine influenza is a contributing factor in chronic respiratory disease problems and the presentation of Porcine Respiratory Disease Complex, a disease syndrome resulting from co-infection with two or more respiratory pathogens. Swine influenza is occasionally associated with fever-induced abortion in sows. IAV infection in pigs involves epithelial cells of the nasal mucosa, tonsils, trachea and lungs. The epithelial cells of the bronchi, bronchioli, and alveoli can be severely compromised during swine influenza infection. An influx of neutrophils and other inflammatory cells leads to epithelial cell necrosis and obstruction of the airways. Equally important is the occurrence of subclinical infections in which swine can become infected with one or more influenza subtypes without overt signs of disease.
2. Natural history of swine influenza viruses around the world
Based on the antigenic differences of the two major surface glycoproteins, hemagglutinin (HA) and neuraminidase (NA), IAVs are classified into subtypes. Currently, H1 and H3 HA subtypes circulate in pigs globally (Fig. 1) associated with different combinations of N1 and N2 NA genes (Vincent et al., 2014a); however, their origin, gene constellation, and antigenic make up vary greatly on different continents (Nelson et al., 2015c). The natural evolution of IAVs is intimately associated with the inherent error prone characteristics of the viral polymerase and the segmented nature of their genome. IAV diversity in pigs is also increased through a combination of mechanisms including gene segment reassortment, point mutations, and introduction of influenza viruses into pigs from other hosts, particularly humans. Once circulating in pigs, this highly dynamic IAV population continues to evolve through further reassortment and antigenic drift continues to shape the evolution of swine IAVs (Nelson et al., 2012a, 2014).
Fig. 1.
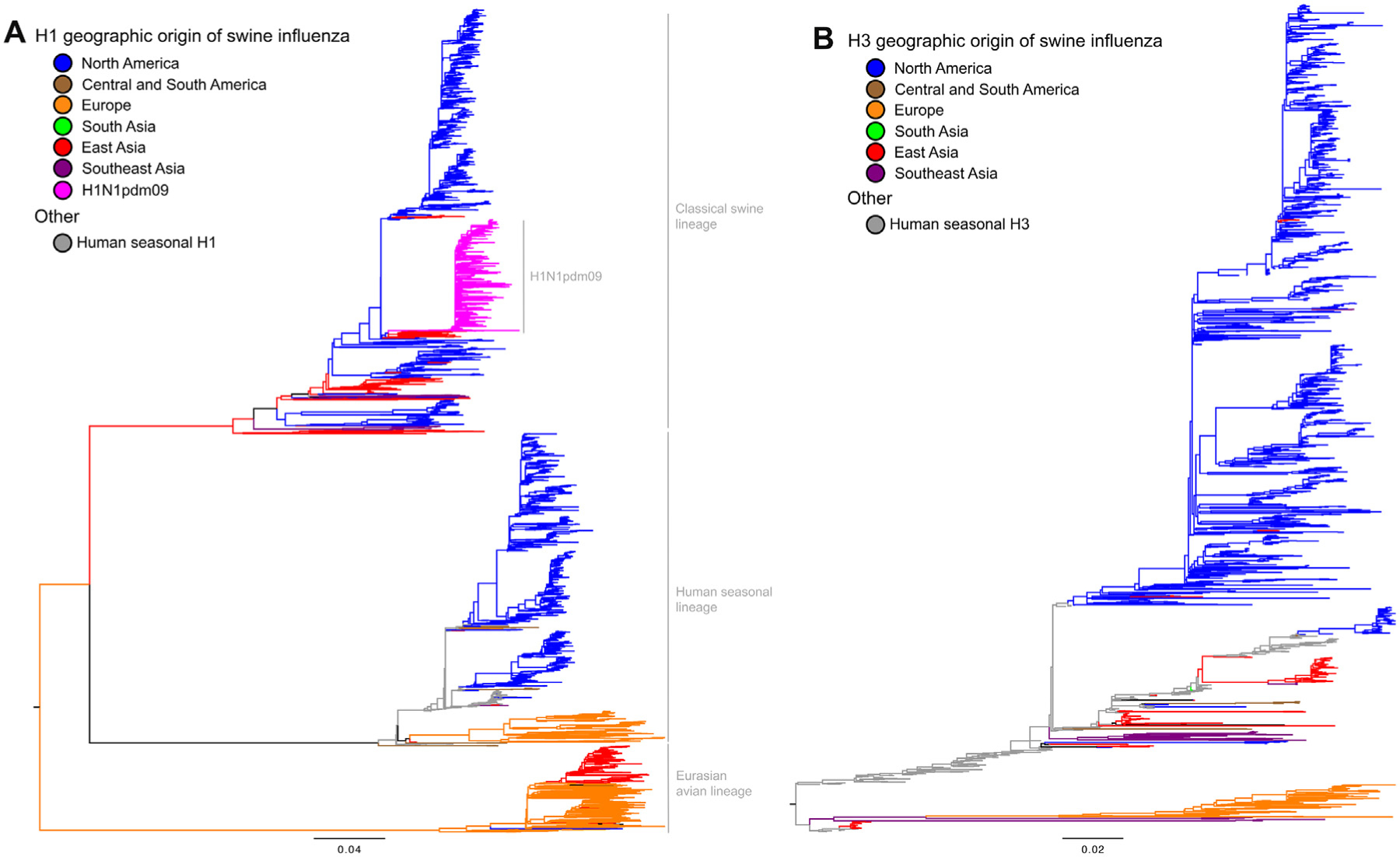
Phylogeny of H1 and H3 influenza A virus hemagglutinin gene sequences from 2000 to present. (A) The best-known tree generated using maximum likelihood methods of 4629 swine and representative human H1 gene sequences is shown: major genetic lineages are labeled, and branch color represents continent of collection. The clade labeled H1N1pdm09 includes HA collected from 34 countries and all continents other than Antarctica. (B) The best-known tree generated using maximum likelihood methods of 1782 swine and representative human H3 gene sequences is shown: branch color represents continent of collection. The geographic groups (data available upon request) include: North America with data from Canada, Mexico, and USA; Central and South America with data from Argentina, Brazil, Chile, Colombia, Costa Rica, Guatemala, and Peru; Europe with data from 12 countries; South Asia with data from Sri Lanka and India; East Asia with data from China, Hong Kong, Japan, South Korea, and Taiwan; and Southeast Asia with data from Singapore, Thailand, and Viet Nam. The trees were midpoint rooted for clarity, all branch lengths are drawn to scale, and the scale bar indicates the number of nucleotide substitutions per site. These analyses used the computational resources of the USDA-ARS computational cluster Ceres on ARS SciNet (www.scinet.usda.ars.gov).
In North America, swine influenza was historically associated with classical swine influenza virus (cs-H1N1) that likely entered into the swine population at the same time of the 1918 H1N1 Spanish flu pandemic. The cs-H1N1 circulated virtually unchanged in this region for almost 80 years until 1998 when swine H3N2 IAVs emerged; reviewed in (Nelson and Vincent, 2015; Vincent et al., 2014b). The new H3N2 viruses were triple reassortant IAVs (tr-H3N2) with HA, NA, and polymerase basic 1 (PB1) gene segments derived from a seasonal human influenza virus, polymerase basic 2 (PB2) and polymerase acidic (PA) gene segments derived from an avian IAV, and nucleoprotein (NP), matrix (M), and nonstructural (NS) gene segments from the classical-swine H1N1. The emergence and perpetuation of swine IAVs carrying the triple reassortant gene (TRIG) cassette changed the ecology of swine influenza not only in North America but also around the world. Further co-circulation of tr-H3N2 and cs-H1N1, as well as additional introductions of HA and/or NA genes from seasonal human IAVs led to the emergence of novel H1N1, H1N2, H3N2 and H3N1 genotypes (Anderson et al., 2015; Lekcharoensuk et al., 2006; Ma et al., 2006; Rajao et al., 2015; Vincent et al., 2008).
Epitomizing the role of swine in the generation of novel genotypes was the 2009 H1N1 pandemic virus (H1N1pdm09), a TRIG swine IAV that incorporated the NA and M gene segments from an Eurasian swine IAV strain and which may have originated in central-west Mexico (Mena et al., 2016; York and Donis, 2013). Although questions remain regarding the emergence of this virus, there is little doubt that the swine-human interface played a major role in this event. Its rapid spread highlights the complexities associated with viral emergence in a dynamic commercial animal production system and the need for robust surveillance in these populations. The spillover of the H1N1pdm09 virus back to pigs in many countries (Nelson et al., 2012a) and the realization that these viruses (or their gene segments) continue to reassort in pigs have renewed interest in worldwide swine surveillance efforts.
In the US, the US Department of Agriculture (USDA) began a surveillance system in 2009 to better characterize the genetic diversity of swine IAVs (Vincent et al., 2014a). There are currently seven different H1 HA genetic clades in the US classified as δ-1, δ-2, α, β, γ, γ2, and pandemic. The γ and δ-1 genetic cluster remain predominant in the US (Vincent et al., 2014b). The H3 viruses are divided into eight different HA genetic clades (IV, IV-A, IV-B, IV-C, IV-D, IV-E, IV-F, and human-like H3s), with clade IV-A representing the majority of circulating H3 in production herds (Kitikoon et al., 2013). The different H1 and H3 genetic lineages currently circulating in the US are most commonly paired with either the N2 NA gene segment from a 2002 human H3N2 seasonal influenza virus or the N1 NA gene segment from the cs-H1N1 virus. A small but consistent proportion of swine isolates contain N2 segments from a 1998 human seasonal lineage virus (Nelson et al., 2012b), mostly paired with δ-2 H1.
The cs-H1N1 virus was detected in Europe until the 1970s, but it was soon replaced in 1979 by the “avian-like” (av-H1N1) swine IAV that continues to persist in Europe. Swine “human-like” H3N2 (hu-H3N2) IAVs also became established in pigs in Europe (Brown, 2013; Zell et al., 2013). The currently circulating hu-H3N2 viruses are reassortants that inherited the internal gene constellation from an av-H1N1 virus in the mid-1980s. In 1994, an H1N2 virus emerged in the UK (hu-H1N2), from a multiple reassortant event involving human seasonal H1N1 from the 1980’s, swine hu-H3N2, and swine av-H1N1 viruses, and spread across Western Europe. Widespread circulation of av-H1N1, hu-H3N2 and hu-H1N2 swine IAVs continues across Europe (Brown, 2013; Zell et al., 2013). The presence of additional reassortant viruses with H1N1pdm09 gene segments has been documented (Simon et al., 2014; Watson et al., 2015), though these appear geographically constrained.
Evolution of swine IAVs in Asia is a result of swine and virus movement as well as human to swine introductions; reviewed in (Choi et al., 2013; Zhu et al., 2013). The cs-H1N1 viruses were recognized as enzootic in China in 1974. Soon after the human H3N2 pandemic (1968 Hong Kong flu), the virus was reported in pigs in several Asian countries where it persisted for few years. Descendants of human H3N2 viruses have repeatedly transmitted to and remained in pigs long after the parental human strains had been replaced in the human population (Zhu et al., 2013). Classical swine H1 viruses that acquired an N2 of contemporary human virus origin also circulated during this time. European H3N2 and H1N1 viruses were first detected in China in 1999 and 2001, respectively, and North American TRIG viruses were found in 2002 (Zhu et al., 2013). In general, many of these viruses are not from a single monophyletic clade, indicating multiple independent introductions. These viruses continued to exchange gene segments, further increasing their diversity. Indeed, H3N2 reassortants with H1N1pdm09-like internal genes and tr-H3N2 surface segments were isolated in Southeast China and are considered potential progenitors of the 2009 H1N1pdm09 virus (Smith et al., 2009). Subsequent studies have characterized additional reassortment events occurring with H1N1pdm09 internal genes, observing additional genotypic diversification (Liang et al., 2014).
Few other Asian countries have had routine surveillance systems in place to monitor swine IAV activity, but the existing information shows regional differences. Swine IAVs that have been detected in South Korea, Japan, Thailand, and Vietnam appear to have unique epidemiologic histories (Choi et al., 2013; Nelson et al., 2015c; Zhu et al., 2013). In South Korea, swine IAVs of North American origin, TRIG reassortants of the H1N1, H1N2, and H3N2 subtypes are common in the pig population; reviewed in (Choi et al., 2013; Zhu et al., 2013). In addition, H3N1 acquired the HA gene from a human seasonal H3N2 virus and other genes from the circulating Korean swine IAVs. In Japan, the cs-H1N1 virus was first reported in 1977 (Choi et al., 2013; Zhu et al., 2013). Additionally, there have been multiple introductions of human H3N2 viruses in swine, but the most commonly reported virus is an H1N2 reassortant that emerged in 1980 with the N2 from a human seasonal H3N2 virus and other gene segments from a cs-H1N1 virus. In Thailand, swine IAVs of North American (mainly cs-H1N1) and Eurasian lineages were detected in the 1980s, along with human lineage H3N2 viruses (Choi et al., 2013; Zhu et al., 2013); multiple reassortment events have led to a diversity of gene constellations. In Vietnam, reassortant H3N2 viruses with the North American TRIG cassette and H3 and N2 segments acquired from 2004 to 2006 human seasonal viruses were reported, along with the parental tr-H3N2 (Baudon et al., 2015; Ngo et al., 2012). Additionally, an H1N2 virus with the N2 and TRIG cassette in association with an H1 from human seasonal 2006 strain was isolated in 2010 (Takemae et al., 2013). H1N1pdm09 viruses have been isolated from pigs in South Korea, Japan, Thailand, and Vietnam: however, a more frequent result of the H1N1pdm09 in this and other regions has been reassortment with cocirculating strains, and a subsequent contribution to internal gene constellation diversity (Lam et al., 2011; Liang et al., 2014; Vijaykrishna et al., 2011).
Following the emergence of the 2009 H1N1pdm09 virus, IAV surveillance in Latin America was also increased; reviewed in (Vincent et al., 2014a). Argentina was the first to report human to pig transmission of the H1N1pdm09 at the peak of the pandemic (Pereda et al., 2011). Since then, the H1N1pdm09 virus has been isolated from pigs in Argentina, Brazil, Chile, Colombia, Costa Rica, Guatemala, Mexico, and Peru. These reports correspond to multiple independent introductions of the H1N1pdm09 virus from humans to pigs and are often associated with respiratory disease in the swine population. In Argentina, Brazil, Chile and Mexico, the emergence of wholly human H3N2 viruses and novel reassortant strains of H1N1 and H1N2 and the H1N1pdm09 virus have been reported, each strain unique to each country or region within the country. In Argentina, there is evidence of circulation of reassortants of H1N1pdm09 and seasonal IAVs: H1N1 and H1N2 viruses with HA and NA segments from (2000–2003) human seasonal H1N1 and H3N2 viruses and H1N1pdm09 internal gene segments were isolated (Pereda et al., 2011). In Brazil, there is evidence of circulation of pdm-H1N1, H1N2, and H3N2 viruses. Phylogenetic analysis revealed independent introductions of human seasonal 1990-like H3N2 and human seasonal 2000-like H1N2 viruses (Nelson et al., 2015b). The pdm-H1N1, and human origin H3N2 and H1N2 viruses have reassorted multiple times in Brazilian pigs with a tendency to maintain internal gene segments from the H1N1pdm09 virus (Nelson et al., 2015b). In Chile, independent introductions of human seasonal viruses, two H3N2s and two H1N1s, have been detected, as well as H1N2 reassortants from these viruses (Nelson et al., 2015a). The swine Chilean H3N2 viruses were probably introduced into pigs in the late 1980s. Likewise, the H1s are most closely related to human seasonal H1N1 viruses from the late 1980s and early 1990s, representing two independent introductions from humans. There is no evidence in Argentina, Brazil, or Chile of swine IAVs of the classical, North American TRIG reassortant or avian-like Eurasian lineages.
3. What to vaccinate against
Understanding both animal and virus movement are needed for the implementation and/or improvement of biosecurity practices or containment measures. Vaccination is another means of mitigating the effects of influenza virus disease and vaccines are most effective if the strains included closely match the currently circulating strains in pigs. Genetic analyses provide panoramic views of the virus landscape at the sequence level in a country or region and thus, can aid in the selection of well-matched swine IAV vaccine strains. A major challenge in selecting appropriate swine IAV vaccine strains is the co-circulation of multiple lineages of viruses in the same region. The HA viral protein is the major target of neutralizing antibodies, and antibodies that inhibit virus hemagglutination of red blood cells as measured in hemagglutination inhibition (HI) assays are considered correlates of protection. Virus variants that are not recognized by previously elicited antibodies can render traditional vaccines that primarily elicit humoral responses ineffective, and therefore result in the need for vaccine strain reformulation and re-vaccination.
For human influenza A and B vaccines, the World Health Organization (WHO) organizes and hosts vaccine strain consultation meetings twice each year to select human vaccine strains for the Northern or Southern Hemispheres (Ampofo et al., 2015). Virologic surveillance is the cornerstone of the WHO vaccine strain selection process, and is carried out through a network of national influenza centers and collaborating centers, with data representing national, regional, and global levels, as well as climatic or transmission zone data aggregation. Laboratory characterization of virus isolates by HI assays remains the gold standard, with specific ferret antisera as well as pre- and post-vaccination human sera. The antigenic data is combined with sequence analysis, virus growth properties, epidemiological, and clinical information to inform the vaccine strain selection process (Russell et al., 2008). Newer technology is being incorporated into these analyses including next generation whole genome sequencing and sequence analysis pipelines, antigenic cartography, and antibody landscape modeling. Improving the understanding of NA evolution and immunity as a consideration for vaccine candidate selection has also been recently re-emphasized. Although these methodologies and concepts can be applied for selection of swine (and other animal) influenza vaccines, a structured system as described in Fig. 2 is still lacking to collect and analyze large sets of viruses from swine at a global scale.
Fig. 2.
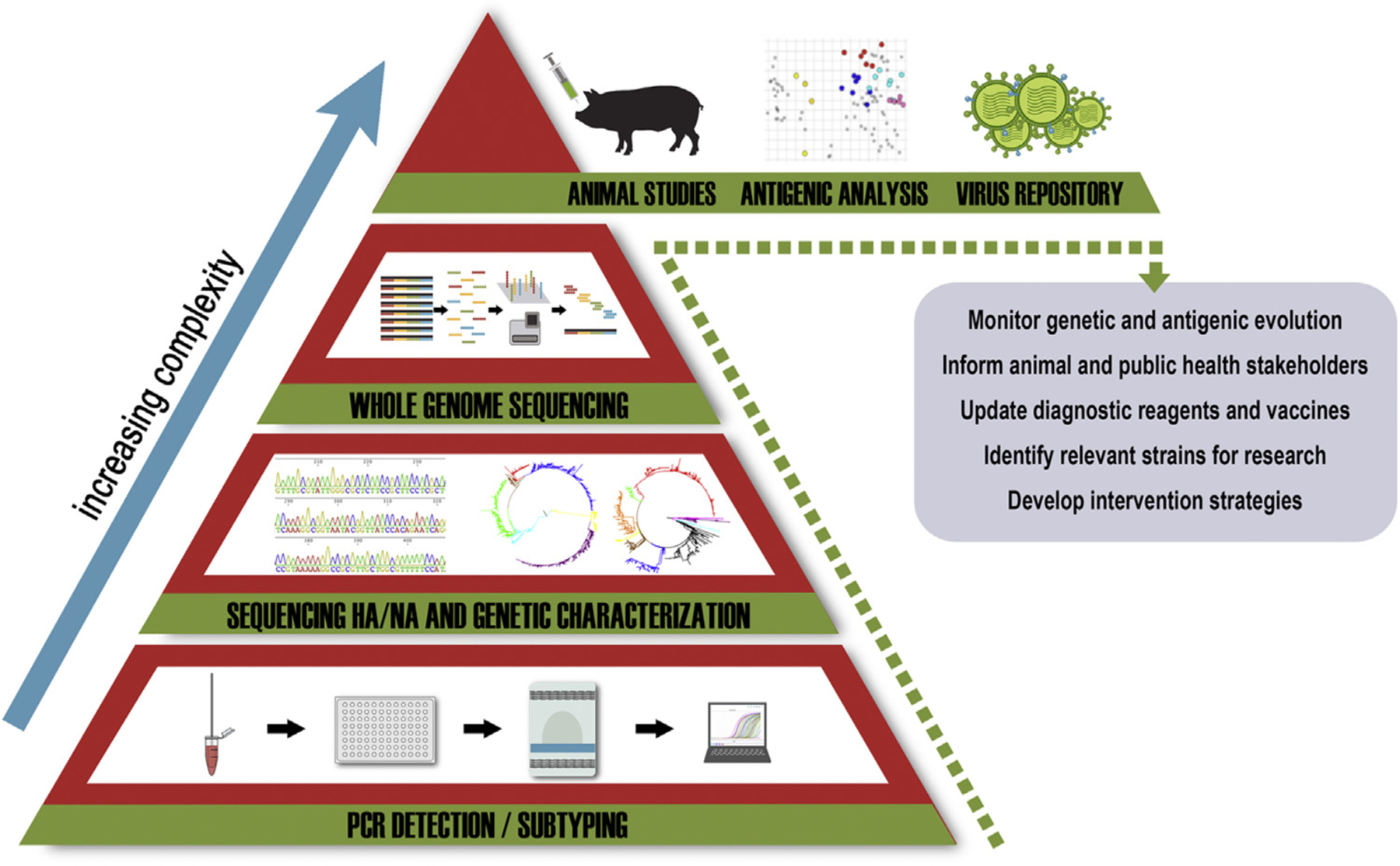
Structure of surveillance for influenza A virus (IAV) in swine. The foundation of a surveillance system is diagnostic screening for presence of IAV. At each level, the number of samples to test are reduced by screening and analyses to identify IAV of interest, but the level of complexity for each subsequent stage increases, allowing for fewer specialized entities to contribute. The ultimate goals of an integrated and collaborative IAV surveillance network are to monitor and report genetic and antigenic evolution to inform animal and public health sectors, update diagnostics and vaccines as needed, identify relevant strains for further research, and to develop and implement improved intervention strategies.
A major advance in tracking IAV evolution and antigenic drift occurred following the deduction of antigenic regions of the 1968 pandemic human H3 virus. These ‘antigenic sites’ have long served as a reference for antigenic positions of relevance to antigenic drift on the globular head of H3, consisting of 131 positions and referred to as regions A-E (Wiley et al., 1981). More recently, antigenic cartography, a computational analysis of antibody-antigen binding assays such as HI assay data (Lapedes and Farber, 2001) has been used not only to characterize the antigenic evolution of human H3 strains but also swine and equine H3 IAV strains (Lewis et al., 2014, 2011; Smith et al., 2004). The distance between viruses in the antigenic map can be used as empirical support for changes in seasonal vaccine strains when there is an 8-fold or greater HI difference between circulating strains and vaccine strain (Cox et al., 2015). This technique was used to identify single amino acid changes that account for phenotypic change. Although more than 130 amino acid positions on the HA were previously characterized as putative antigenic sites, major substitutions leading to the emergence of antigenically distinct seasonal epidemic viruses occurred at just 7 amino acid positions in the HA of the human seasonal H3 (Koel et al., 2013). The H3N2 component of the human vaccine for 2015–2016 was recommended to be changed based on a change to a single amino acid at position 159, serving as a recent example that a single amino acid substitution can be associated with a loss in vaccine efficacy.
Analyses of the antigenic evolution of swine H3 field isolates from the United States identified two major antigenic clusters and several distinct outlier groups or strains (Lewis et al., 2014). Six of the 7 positions previously identified in human seasonal H3 (positions 145, 155, 156, 158, 159, 189, and 193) were also associated with cluster/outlier transition in the swine H3 wild type viruses (Lewis et al., 2014). Abente et al. quantified the effect of mutations at these six positions using reverse genetics, and in vitro and in vivo experiments (Abente et al., 2016). Typically, at least two amino acid changes were required to impart a significant antigenic change but these studies were restricted to a subset of field isolates, and it is possible that single amino acid mutations would have major antigenic effects as described for human strains under field conditions.
The antigenic structure of the H1 HA was also characterized, in which four immunodominant antigenic regions or sites were determined, designated Sa, Sb, Ca, Cb (Caton et al., 1982). Major antigenic changes in the recent evolution of seasonal H1N1 viruses (pre-H1N1pdm09) were predominantly caused by single amino acid substitutions near the receptor-binding site in previously determined antigenic sites (Koel et al., 2013, 2015). The H1N1pdm09 viruses have not yet undergone a major antigenic transition; however, substitutions in or near the receptor-binding site (mostly in the 151–159 loop) were shown to influence the antigenic properties of these viruses (Koel et al., 2015). Genetic diversity greatly influences the antigenic diversity of currently circulating U.S. swine H1 IAV; however, antigenic changes have not been correlated with single substitutions at specific amino acid positions at this time. More recently, Lewis et al. quantified the antigenic diversity of swine influenza viruses on a multi-continental scale using the largest set of swine influenza virus antigenic data compiled to date (Lewis et al., 2016). Hundreds of H1 and H3 strains were analyzed (Fig. 3). Significant antigenic variations were observed particularly as the result of multiple cross-over events of human influenza viruses into pigs and subsequent perpetuation of these viruses in the pig population. At the core of these analyses is the realization that HA and NA antigen selections for vaccine components for swine influenza vaccines is far more complex than for humans and will require decisions at the country or regional levels, perhaps at even finer-scaled levels.
Fig. 3.
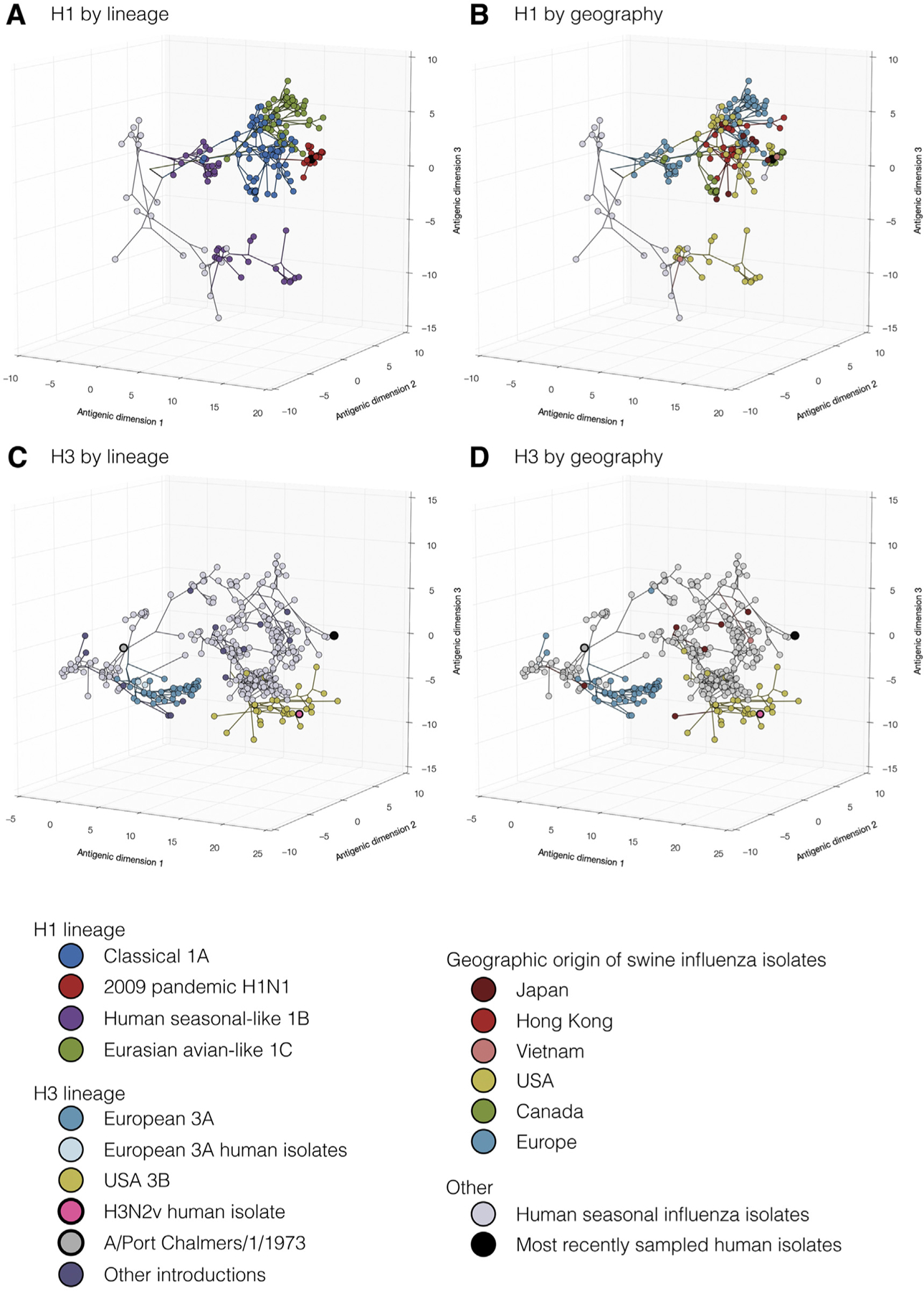
Evolutionary relationships of H1 (A, B) and H3 (C, D) influenza viruses circulating in swine and humans inferred by Bayesian Multi-dimensional scaling (BMDS). Each colored ball represents a single virus. Viruses are colored by lineage (A,C) and by geography (B,D). Lines connecting each virus represent inferred phylogenetic relationships. Distances for antigenic dimensions are measured in antigenic units (AU) and each unit is equivalent to a two-fold dilution in HI assay data. Viruses close to one another are more antigenically similar than viruses further apart. Reprinted from Lewis et al., 2016; doi:10.7554/eLife.12217.003.
3.1. What to vaccinate with
Not all countries with actively circulating swine influenza viruses use vaccines as a means to control disease. It is expected, however, that as swine production intensifies worldwide, more countries will rely on the use of vaccines to control swine influenza. Previously, all the influenza vaccines licensed in the United States were whole inactivated virus (WIV) products typically combined with potent oil-in-water adjuvants (Fig. 4). Adjuvanted WIV vaccines usually stimulate robust antibody responses that can be measured by HI assays. Different adjuvants can alter the response to antigens by stimulating different arms of the immune system or stimulating broader cross-reactive antibodies. However, experimental data suggest that the protection provided by commercial WIV against contemporary IAV is limited, in part due to the diversity of viruses we describe above; reviewed in (Van Reeth and Ma, 2013). In an attempt to overcome this challenge, the USDA Center for Veterinary Biologics implemented a new licensure policy in 2007 that allows vaccine manufacturers with current licensing to update vaccine strains to reflect contemporary genetic diversity. Further, in 2012, a new vaccine platform product was licensed for swine H3N2 virus (Harrisvaccines, 2012) using technology based on a non-replicating alphavirus RNA particle (Vander Veen et al., 2012, 2013). These approaches are used alongside autogenous vaccines which account for approximately 50% of vaccines produced in 2008 to 2011. A recent review by Sandbulte et al. provided guidance for choosing vaccines based on the currently available technology (Sandbulte et al., 2015).
Fig. 4.
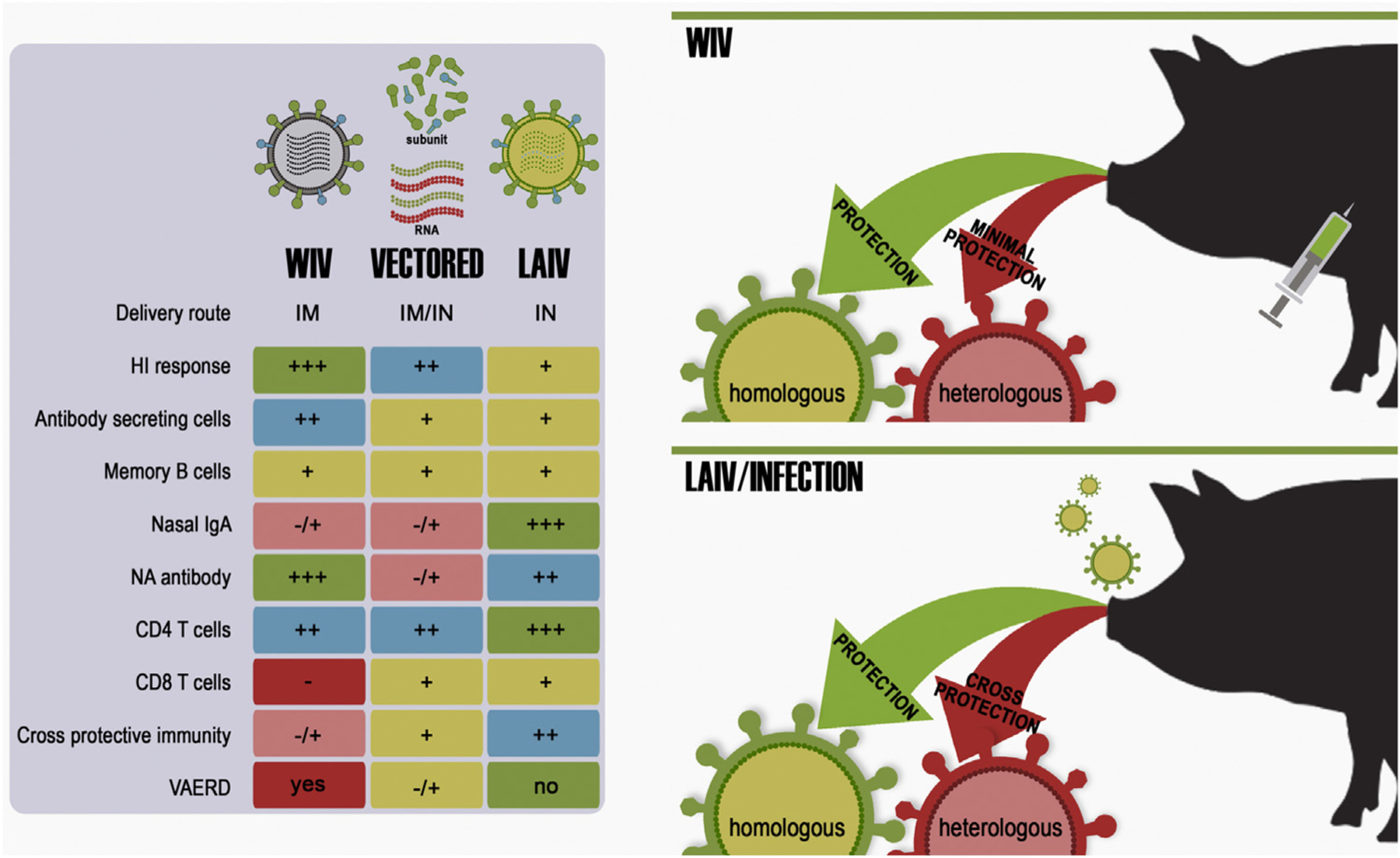
Vaccine platform and route of administration influence the resulting immune response. Whole inactivated vaccines (WIV) are administered parenterally through intramuscular routes whereas live attenuated influenza vaccines (LAIV) are typically delivered mucosally through intranasal routes. Vectored or RNA vaccines may be delivered through either route. The route of administration as well as the manner in which vaccine antigens are presented to the immune system influences the resulting host immune response.
Licensed products must be demonstrated to be efficacious in experimental studies with homologous or related viruses, yet they frequently fail or perform sub-optimally in the context of field application. Manufacturers are not required to inform or include strain names of IAV contained in the vaccine on the label, so we cannot speculate on the potential coverage of commercial vaccines. These factors complicate our understanding of which antigens are being routinely used for vaccination and obscure large-scale objective measures. In contrast to human IAV vaccine efficacy evaluation, there is no continued evaluation by regulatory authorities on efficacy of licensed swine vaccines in the field following licensure.
3.2. Why vaccines fail
Although sterilizing immunity would be ideal, the reality is that this type of vaccine immunity is rarely achievable with IAV, at least long term, due to its remarkable ability to evolve and evade host immunity. The more realistic goals are to prevent clinical disease and reduce virus replication and shedding to the extent that transmission events are greatly reduced without causing immunopathology. While vaccination with WIV induces robust HI antibody responses, protection depends on the vaccine and challenge virus being closely related, and often only results in partial protection (Kitikoon et al., 2006; Loving et al., 2013). Further, absence of serum HI titers does not always indicate lack of protection if cell-mediated immune and mucosal responses are stimulated, as it has been shown after the use of live attenuated influenza vaccines (LAIV) in pigs (Vincent et al., 2012). Although traditional IAV vaccines seem fairly basic, there are many mitigating factors that can interfere with IAV vaccines being successful at achieving these goals. At a basic level, the importance of antigenic balance in multivalent vaccines, especially with H3 and H1 components, is well known (Quinnan et al., 1983). Some viruses are poor antigens and induce minimal HI titers even with adjuvants. When implementing a new vaccine formulation or strategy into a herd, pre- and post-vaccination serum should be tested against each of the vaccine antigens in the HI assay at the minimum. HI tests with post-vaccination serum against herd-specific outbreak strains gives further confidence in the vaccine coverage. In addition to considerations with vaccine immunogenicity, WIV used in the field can have diminished performance, even with vaccines that performed well under test conditions due to more complicated host immune factors.
Monitoring immunologic profiles in individuals with a series of IAV exposures over time revealed potentially complex immune responses to conserved and heterologous epitopes of IAV. The sequence of exposures is extremely important to each subsequent exposure, especially with vaccines. The concept of original antigenic sin with IAV (Fazekas de St and Webster, 1966), whereby the strain specific response to the first exposure blocked a strain specific response to a second virus of a distant antigenic variant of the same subtype or a different subtype, has had major implications in the development of influenza vaccines. Generally, the second heterologous exposure boosted the antibody response to the initial priming exposure at the expense of the secondary response. This has also been shown to happen with less extreme pairs of heterologous viruses, where the heterologous secondary exposure still boosts the priming exposure, but the secondary exposure is only muted rather than blocked, a phenomenon known as back-boosting (Fonville et al., 2016). Additionally, the initial infection with IAV that an individual experiences has been proposed to shape every subsequent response, termed antigenic imprinting (Worobey et al., 2014), and may have impacted mortality patterns during pandemics, particularly the 1918 pandemic.
An additional complicating factor has recently been identified in pigs, termed vaccine- associated enhanced respiratory disease (VAERD). VAERD occurs when pigs are vaccinated with oil-in-water adjuvanted WIV and challenged with a homosubtypic but antigenically distinct IAV and results in extensive lung damage. The immune response elicited from the vaccination consists of cross-reactive antibodies that fail to neutralize the mismatched virus (Gauger et al., 2011; Rajao et al., 2014). Understanding these phenomena in pigs at the individual and population levels may be used to avoid unwanted responses and even leveraged to enhance protection, such as the potential for heterologous prime and boost vaccine strategies to broaden the antibody repertoire or intentionally inducing a back-boost with a heterologous boost vaccine to increase the antibody titers to the priming exposure.
Control of IAV infection in swine production systems traditionally relied on the vaccination of breeding females with WIV to result in the passive transfer of antibodies to their piglets via colostrum. Ideally, the titers of the maternally derived antibodies (MDA) should be high enough to protect the piglets during the first weeks of life before they can mount a protective active immune response. However, these passive antibodies can interfere with active humoral and cellular immune responses to vaccine and may lead to VAERD when pigs are vaccinated in the presence of MDA and then exposed to heterologous virus (Loeffen et al., 2003; Vincent et al., 2012). MDA do not usually protect piglets against infection and transmission but rather against clinical disease, provided the antibodies match the IAV strain to which pigs are exposed (Allerson et al., 2013, 2014). Mismatched MDA alone was shown to negatively impact the outcome of disease as MDA-positive pigs infected with heterologous virus developed VAERD (Rajao et al., 2016). These complicated immune interactions with IAV are important to keep in perspective when considering the great diversity of IAV circulating in pigs currently and the widespread use of inactivated products for immunization of pigs.
3.3. The way forward
Biological and computational methods to better understand and quantify the antigenic evolution of influenza viruses, together with relevant correlates of protection, are greatly needed to inform the decision-making process for vaccine strain selection. On the vaccine front, several approaches are under development and have shown great promise against swine influenza. Subunit and/or vectored (baculovirus- adenovirus-, plasmids-based) vaccines targeting the HA, NA, and/or the more conserved M or NP proteins have also demonstrated efficacy against IAV in swine (Braucher et al., 2012; Wesley and Lager, 2006), and some have clinical or manufacturing advantages over WIV vaccines. There is renewed interest in messenger RNA (mRNA)-based vaccines due to significant improvements in methods and reagents to increase mRNA stability, delivery, and activity, including TLR7-mediated effects (Petsch et al., 2012). mRNA-based vaccines can be easily delivered intramuscularly or intradermally where innate pattern-recognition receptors in target dendritic cells take up the mRNA, synthesize the encoded product and stimulate protective humoral and CD8+ responses. There are many advantages of mRNA-based vaccines over other recombinant approaches: it is non-replicating, is a fully defined genetic vector with limited persistence and antigen expression, has no possibility for recombination, and allows for precise control over pharmacokinetics and dosing, thus potentially reducing the need for adjuvants. In addition, the production of mRNA vaccines is flexible and highly scalable and, for influenza vaccines, it does not require growing virus in eggs. Studies by Petsch, et. al., showed that naive pigs immunized with triple mRNA vaccine formulation encoding HA, NA, and NP were protected from mild disease and vaccinated pigs showed reduced virus titers after challenge (Petsch et al., 2012). Despite many potential advantages, recombinant, DNA-based or RNA-based subunit vaccine strategies have to be meticulously designed to contain contemporary antigens to increase antigenic coverage of the circulating viral diversity. Furthermore, it remains to be determined whether these alternative approaches can overcome VAERD as observed with currently licensed vaccines.
The concept of developing vaccines that induce broadly cross-protective and long lasting immunity has been a focus for human IAV research for many years (Cox et al., 2015). The term “universal” vaccine was coined from this field of study. Universal vaccine platforms include viral-like particles expressing multiple HA and NA subtypes, vectored peptides from highly conserved epitopes of HA or NA, as well as several strategies targeting the more conserved HA2 stem or stalk (Wiersma et al., 2015). One stem-targeting approach is to use sequential heterologous exposures to vaccine viruses with chimeric HA proteins (Krammer et al., 2014; Nachbagauer et al., 2014). The HA1 globular head containing the highly variant portion of the mature HA is replaced by HA1 from an irrelevant subtype. Each series of the chimeric vaccines contain the conserved HA2 stem from a relevant IAV strain to drive a more robust antibody response to the conserved HA2 stem. Although antibodies induced from this platform are not neutralizing in the traditional sense, this vaccine strategy can reduce viral shedding and transmission and prevent clinical disease against heterosubtypic viruses with the conserved stem in laboratory animal models. In pigs, however, one study revealed that anti-stalk antibodies generated by WIV were associated with VAERD (Khurana et al., 2013). Thus, a universal influenza vaccine in pigs will need to be carefully evaluated to avoid eliciting antibodies with detrimental features or outcomes.
Research in swine on LAIV has repeatedly demonstrated their general safety and superior efficacy under experimental conditions that are more similar to those in field-settings when directly compared to inactivated vaccines (Fig. 4) (Masic et al., 2009; Pena et al., 2011; Richt et al., 2006; Solorzano et al., 2005; Vincent et al., 2007). Experimental data demonstrated that attenuated vaccines are superior in their ability to overcome MDA interference, protect against heterologous antigenically distinct viruses, as well as protect against transmission of heterologous challenge viruses to naïve contact pigs (Kappes et al., 2012; Loving et al., 2013). More importantly, LAIVs have not been associated with VAERD under experimental conditions. Three types of LAIV platforms have been tested in swine: (1) a replication defective swine influenza virus carrying a HA gene segment encoding a cleavage site recognized by elastase (eHA), instead of the natural amino acid sequence recognized by trypsin-like proteases; (2) viruses based on various deletions of the viral NS1 ORF (ΔNS1); and (3) a two-combination strategy with temperature sensitive amino acid mutations in the PB2 and PB1 segments and addition of a HA tag at the C-terminus of PB1 (tsHAtag). Elastase is produced by neutrophils but is not typically released into the lumen of the respiratory tract. Thus, the eHA LAIV virus displays limited replication in pigs. Pigs vaccinated intratracheally with the eHA LAIV virus were protected after swine IAV challenge by showing reduced lung pathology and reduced virus titers. Likewise, LAIVs based on either the ΔNS1 or the tsHAtag have been shown safe after intranasal delivery and highly protective in various challenge studies. Both of these LAIV vaccines have shown significant cross-protection after challenge with antigenically distinct viruses but within the same subtype. In a side-by-side study, various commercially available WIV vaccines were compared to the ΔNS1 and tsHAtag LAIVs in terms of protective efficacy against a cluster IV H3N2 swine IAV (Loving et al., 2013). In the same study, the capacity of transmission of the challenge virus from vaccinated pigs to naïve pigs was analyzed. Low levels of virus shedding after challenge from vaccinated pigs were sufficient to initiate a round of virus transmission among naïve pigs. The only group showing sterilizing immunity, and therefore no transmission to naïve pigs, was vaccinated with the tsHAtag LAIV. Lack of cell mediated responses in the WIV groups may explain their lower performance compared to the LAIV groups. Greater antigenic differences between the ΔNS1 LAIV (cluster I) and the tsHAtg LAIV (cluster IV) compared to the challenge virus (cluster IV) may have accounted for lack of sterilizing immunity in the former. This and other studies, however, revealed that the caveat to the LAIV are the limitations of the HI assay in predicting protection or cross-protection. Alternative, more refined, methods to measure protective responses using LAIV are greatly needed. In our studies, LAIVs have consistently provided evidence of higher IgA responses in lung lavages. From a more practical perspective, increase IgA levels in pig serum is usually detected after LAIV; however, it remains to be established the levels of IgA that correlate with protection. In large swine production settings, intranasal delivery of LAIVs is perhaps more cumbersome than intramuscular WIV administration.
The risk of reassortment of LAIVs with wild type swine strains is perhaps very low but it cannot be overlooked. If LAIVs become mainstream for the prevention and control of swine influenza in the field, it will require close monitoring as their misuse could create more problems than they solve. Mixing of different vaccines from different sources is not uncommon in the field. The rationale behind this type of practice is to increase the chances of protection against disease while minimizing the number of times a pig needs to be restrained or handled for immunizations. If, for example, a ΔNS1 LAIV vaccine were to be mixed with a eHA LAIV or tsHAtag LAIV vaccine prior to administration to pigs, there is a chance that a wild type swine IAV will be generated with gene segments derived from each of unmodified gene segments in each LAIV vaccine. Such scenarios may lead to even more diversity of swine IAV strains or strains with increased zoonotic potential. Incorporation of the HA segment into a wild type backbone is the most significant risk from animal and public health perspectives. Thus, it would perhaps be advisable to consider the incorporation of molecular markers in the HA segment that would reduce its fitness and capacity to reassort and allow for easy detection and differentiation from wild type segments.
4. Conclusions
The key questions are how can we use current vaccines to improve the ability to control IAV in swine and what is on the horizon in the future? The first question requires going back to basic epidemiology and disease control. Each farm needs to know the virus status and immune profile at the pig level and in different age groups. Each pig production flow needs to know the virus status and population immune profiles at farm and flow levels. When do MDA wane and does hyper-immunizing sows with WIV negate the use of vaccine in growing pigs? What are the consequences for IAV evolution and emergence of novel variants within the pig flow with this strategy? What are the consequences if our current sow vaccination strategies were changed? What are the IAVs circulating at the regional level, i.e., what IAV do your neighbors and pig sources have? How are IAV transmitted, transported, or otherwise shared within pig production flows and between regions and farms? Once these profiles are established, endemic and outbreak strains need to be matched to vaccines. This requires knowledge at the genetic and antigenic levels for the strains contained in the vaccines. Can different vaccination strategies be employed to increase effectiveness by expanding the breadth or quality of immunity? Different vaccine application strategies should be tested in swine to better understand the impact of heterologous prime-boost or the effects of applying different vaccines at different timings based on immune profiles as discussed above.
In the future, new vaccine platforms may be on the market soon that will provide alternative options to those currently available. Nonetheless, an integrated multi-agency approach is needed to improve IAV vaccine strain selection for use in swine. The USDA IAV in swine surveillance system has reached a critical mass and is in a position to provide input to national and regional vaccine strain selection, but surveillance alone does not lead to better vaccines. Finer scale resolution of spatio-temporal trends in subtype, genotype, and antigenic diversity will allow the identification of long-term patterns of transmission and evolution. Representative viruses from predominant phylogenetic clusters should be evaluated at an antigenic level for serologic cross-reactivity with anti-sera from vaccinated swine. Additional swine antisera should be generated with IAV that have drifted antigenically or with newly added vaccine strains. In vivo swine studies should be conducted when field strains of IAV with substantial changes have been identified or when vaccine strains have been substantially changed. Following the human vaccine model, IAV backbones could be approved by the Center for Veterinary Biologics (CVB) to permit timely update with new and relevant HA and NA for vaccine seed strains. These backbones should exhibit high yield growth properties as well as attenuating mutations in the case of LAIV. Companies may select viruses based on HA and NA sequences from the surveillance system or from their own internal surveillance data for their customers. Viruses from the USDA IAV repository are readily available for this purpose.
Continued funding for surveillance and research, greater collaboration, and translation of newer methodologies from human and other host species for use in swine are required in order to improve control strategies to better mitigate the effects of continuous circulation of diverse swine IAV. Greater transparency with farm and flow level meta-data to connect with virus sequence data is needed to fully understand virus evolution and migration and build upon the foundation established with the surveillance system thus far. Only then can vaccine strains and platforms be better matched to meet the needs of the swine industry.
Acknowledgements
We gratefully acknowledge the pork producers, swine veterinarians, and laboratories for participating in the USDA IAV surveillance system in swine and other surveillance activities around the globe, establishing the critical foundation from which to build future strategies to prevent and control IAV in swine. Authors were supported by USDA-ARS, USDA-APHIS, or by an NIH-National Institute of Allergy and Infectious Diseases (NIAID) interagency agreement (R21AI098079) and the Center of Research in Influenza Pathogenesis, an NIAID funded Center of Excellence in Influenza Research and Surveillance (CEIRS, HHSN272201400008C). NSL was supported in part by USDA-ARS agreement (58-3625-4-071-F). EJA, RRW, and TKA have appointments with the ARS-USDA Research Participation Program administered by the Oak Ridge Institute for Science and Education (ORISE) through an interagency agreement between the U.S. Department of Energy (DOE) and USDA. ORISE is managed by ORAU under DOE contract number DE-AC05-06OR23100. The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication. Mention of trade names or commercial products in this article is solely for the purpose of providing specific information and does not imply recommendation or endorsement by the USDA. USDA is an equal opportunity provider and employer.
Footnotes
Conflicts of interest
The authors declare that there are no conflicts of interest.
References
- Abente EJ, Santos J, Lewis NS, Gauger PC, Stratton J, Skepner E, Anderson TK, Rajao DS, Perez DR, Vincent AL, 2016. The molecular determinants of antibody recognition and antigenic drift in the H3 hemagglutinin of swine influenza A virus. J. Virol 90, 8266–8280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allerson M, Deen J, Detmer SE, Gramer MR, Joo HS, Romagosa A, Torremorell M, 2013. The impact of maternally derived immunity on influenza A virus transmission in neonatal pig populations. Vaccine 31, 500–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allerson MW, Davies PR, Gramer MR, Torremorell M, 2014. Infection dynamics of pandemic 2009 H1N1 influenza virus in a two-site swine herd. Transbound. Emerg. Dis 61, 490–499. [DOI] [PubMed] [Google Scholar]
- Ampofo WK, Azziz-Baumgartner E, Bashir U, Cox NJ, Fasce R, Giovanni M, Grohmann G, Huang S, Katz J, Mironenko A, Mokhtari-Azad T, Sasono PM, Rahman M, Sawanpanyalert P, Siqueira M, Waddell AL, Waiboci L, Wood J, Zhang W, Ziegler T, Group WHOW, 2015. Strengthening the influenza vaccine virus selection and development process: report of the 3rd WHO informal consultation for improving influenza vaccine virus selection held at WHO headquarters, Geneva, Switzerland, 1–3 april 2014. Vaccine 33, 4368–4382. [DOI] [PubMed] [Google Scholar]
- Anderson TK, Campbell BA, Nelson MI, Lewis NS, Janas-Martindale A, Killian ML, Vincent AL, 2015. Characterization of co-circulating swine influenza A viruses in North America and the identification of a novel H1 genetic clade with antigenic significance. Virus Res. 201, 24–31. [DOI] [PubMed] [Google Scholar]
- Baudon E, Poon LL, Dao TD, Pham NT, Cowling BJ, Peyre M, Nguyen KV, Peiris M, 2015. Detection of novel reassortant influenza a (H3N2) and H1N1 2009 pandemic viruses in swine in hanoi, vietnam. Zoonoses Public Health 62, 429–434. [DOI] [PubMed] [Google Scholar]
- Braucher DR, Henningson JN, Loving CL, Vincent AL, Kim E, Steitz J, Gambotto AA, Kehrli ME Jr., 2012. Intranasal vaccination with replication-defective adenovirus type 5 encoding influenza virus hemagglutinin elicits protective immunity to homologous challenge and partial protection to heterologous challenge in pigs. Clin. Vaccine Immunol: CVI 19, 1722–1729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown IH, 2013. History and epidemiology of Swine influenza in Europe. Curr. Top. Microbiol. Immunol 370, 133–146. [DOI] [PubMed] [Google Scholar]
- Caton AJ, Brownlee GG, Yewdell JW, Gerhard W, 1982. The antigenic structure of the influenza virus A/PR/8/34 hemagglutinin (H1 subtype). Cell 31, 417–427. [DOI] [PubMed] [Google Scholar]
- Choi YK, Pascua PN, Song MS, 2013. Swine influenza viruses: an Asian perspective. Curr. Top. Microbiol. Immunol 370, 147–172. [DOI] [PubMed] [Google Scholar]
- Cox NJ, Hickling J, Jones R, Rimmelzwaan GF, Lambert LC, Boslego J, Rudenko L, Yeolekar L, Robertson JS, Hombach J, Ortiz JR, 2015. Report on the second WHO integrated meeting on development and clinical trials of influenza vaccines that induce broadly protective and long-lasting immune responses: geneva, Switzerland, 5–7 May 2014. Vaccine 33, 6503–6510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fazekas de St G, Webster RG, 1966. Disquisitions of original antigenic sin. I. Evidence in man. J. Exp. Med 124, 331–345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fonville JM, Fraaij PL, de Mutsert G, Wilks SH, van Beek R, Fouchier RA, Rimmelzwaan GF, 2016. Antigenic maps of influenza A(H3N2) produced with human antisera obtained after primary infection. J. Infect. Dis 213, 31–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gauger PC, Vincent AL, Loving CL, Lager KM, Janke BH, Kehrli ME Jr., Roth JA, 2011. Enhanced pneumonia and disease in pigs vaccinated with an inactivated human-like (delta-cluster) H1N2 vaccine and challenged with pandemic 2009 H1N1 influenza virus. Vaccine 29, 2712–2719. [DOI] [PubMed] [Google Scholar]
- Harrisvaccines 2012. Harrisvaccines awarded vaccine product licensure
- Kappes MA, Sandbulte MR, Platt R, Wang C, Lager KM, Henningson JN, Lorusso A, Vincent AL, Loving CL, Roth JA, Kehrli ME Jr., 2012. Vaccination with NS1-truncated H3N2 swine influenza virus primes T cells and confers cross-protection against an H1N1 heterosubtypic challenge in pigs. Vaccine 30, 280–288. [DOI] [PubMed] [Google Scholar]
- Khurana S, Loving CL, Manischewitz J, King LR, Gauger PC, Henningson J, Vincent AL, Golding H, 2013. Vaccine-induced anti-HA2 antibodies promote virus fusion and enhance influenza virus respiratory disease. Sci. Transl. Med 5, 200ra114. [DOI] [PubMed] [Google Scholar]
- Kitikoon P, Nilubol D, Erickson BJ, Janke BH, Hoover T, Sornsen S, Thacker EL, 2006. The immune response and maternal antibody interference to a heterologous H1N1 swine influenza virus infection following vaccination. Vet. Immunol. Immunopathol 112, 117–128. [DOI] [PubMed] [Google Scholar]
- Kitikoon P, Nelson MI, Killian ML, Anderson TK, Koster L, Culhane MR, Vincent AL, 2013. Genotype patterns of contemporary reassorted H3N2 virus in US swine. J. Gen. Virol 94, 1236–1241. [DOI] [PubMed] [Google Scholar]
- Koel BF, Burke DF, Bestebroer TM, van der Vliet S, Zondag GC, Vervaet G, Skepner E, Lewis NS, Spronken MI, Russell CA, Eropkin MY, Hurt AC, Barr IG, de Jong JC, Rimmelzwaan GF, Osterhaus AD, Fouchier RA, Smith DJ, 2013. Substitutions near the receptor binding site determine major antigenic change during influenza virus evolution. Science 342, 976–979. [DOI] [PubMed] [Google Scholar]
- Koel BF, Mogling R, Chutinimitkul S, Fraaij PL, Burke DF, van der Vliet S, de Wit E, Bestebroer TM, Rimmelzwaan GF, Osterhaus AD, Smith DJ, Fouchier RA, de Graaf M, 2015. Identification of amino acid substitutions supporting antigenic change of influenza A(H1N1)pdm09 viruses. J. Virol 89, 3763–3775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krammer F, Hai R, Yondola M, Tan GS, Leyva-Grado VH, Ryder AB, Miller MS, Rose JK, Palese P, Garcia-Sastre A, Albrecht RA, 2014. Assessment of influenza virus hemagglutinin stalk-based immunity in ferrets. J. Virol 88, 3432–3442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam TT, Zhu H, Wang J, Smith DK, Holmes EC, Webster RG, Webby R, Peiris JM, Guan Y, 2011. Reassortment events among swine influenza A viruses in China: implications for the origin of the 2009 influenza pandemic. J. Virol 85, 10279–10285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lapedes A, Farber R, 2001. The geometry of shape space: application to influenza. J. Theor. Biol 212, 57–69. [DOI] [PubMed] [Google Scholar]
- Lekcharoensuk P, Lager KM, Vemulapalli R, Woodruff M, Vincent AL, Richt JA, 2006. Novel swine influenza virus subtype H3N1, United States. Emerg. Infect. Dis 12, 787–794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis NS, Daly JM, Russell CA, Horton DL, Skepner E, Bryant NA, Burke DF, Rash AS, Wood JL, Chambers TM, Fouchier RA, Mumford JA, Elton DM, Smith DJ, 2011. Antigenic and genetic evolution of equine influenza A (H3N8) virus from 1968 to 2007. J. Virol 85, 12742–12749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis NS, Anderson TK, Kitikoon P, Skepner E, Burke DF, Vincent AL, 2014. Substitutions near the hemagglutinin receptor-binding site determine the antigenic evolution of influenza A H3N2 viruses in U.S swine. J. Virol 88, 4752–4763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis NS, Russell CA, Langat P, Anderson TK, Berger K, Bielejec F, Burke DF, Dudas G, Fonville JM, Fouchier RA, Kellam P, Koel BF, Lemey P, Nguyen T, Nuansrichy B, Peiris JM, Saito T, Simon G, Skepner E, Takemae N, consortium E, Webby RJ, Van Reeth K, Brookes SM, Larsen L, Watson SJ, Brown IH, Vincent AL, 2016. The global antigenic diversity of swine influenza A viruses. Elife 5, e12217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang H, Lam TT, Fan X, Chen X, Zeng Y, Zhou J, Duan L, Tse M, Chan CH, Li L, Leung TY, Yip CH, Cheung CL, Zhou B, Smith DK, Poon LL, Peiris M, Guan Y, Zhu H, 2014. Expansion of genotypic diversity and establishment of 2009 H1N1 pandemic-origin internal genes in pigs in China. J. Virol 88, 10864–10874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loeffen WL, Heinen PP, Bianchi AT, Hunneman WA, Verheijden JH, 2003. Effect of maternally derived antibodies on the clinical signs and immune response in pigs after primary and secondary infection with an influenza H1N1 virus. Vet. Immunol. Immunopathol 92, 23–35. [DOI] [PubMed] [Google Scholar]
- Loving CL, Lager KM, Vincent AL, Brockmeier SL, Gauger PC, Anderson TK, Kitikoon P, Perez DR, Kehrli ME Jr., 2013. Efficacy in pigs of inactivated and live attenuated influenza virus vaccines against infection and transmission of an emerging H3N2 similar to the 2011–2012 H3N2 v. J. Virol 87, 9895–9903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma W, Gramer M, Rossow K, Yoon KJ, 2006. Isolation and genetic characterization of new reassortant H3N1 swine influenza virus from pigs in the midwestern United States. J. Virol 80, 5092–5096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masic A, Booth JS, Mutwiri GK, Babiuk LA, Zhou Y, 2009. Elastase-dependent live attenuated swine influenza A viruses are immunogenic and confer protection against swine influenza A virus infection in pigs. J. Virol 83, 10198–10210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mena I, Nelson MI, Quezada-Monroy F, Dutta J, Cortes-Fernandez R, LaraPuente JH, Castro-Peralta F, Cunha LF, Trovao NS, Lozano-Dubernard B, Rambaut A, van Bakel H, Garcia-Sastre A, 2016. Origins of the 2009 H1N1 influenza pandemic in swine in Mexico. Elife 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nachbagauer R, Wohlbold TJ, Hirsh A, Hai R, Sjursen H, Palese P, Cox RJ, Krammer F, 2014. Induction of broadly reactive anti-hemagglutinin stalk antibodies by an H5N1 vaccine in humans. J. Virol 88, 13260–13268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson MI, Vincent AL, 2015. Reverse zoonosis of influenza to swine: new perspectives on the human-animal interface. Trends Microbiol. 23, 142–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson MI, Gramer MR, Vincent AL, Holmes EC, 2012a. Global transmission of influenza viruses from humans to swine. J. Gen. Virol 93, 2195–2203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson MI, Vincent AL, Kitikoon P, Holmes EC, Gramer MR, 2012b. Evolution of novel reassortant A/H3N2 influenza A viruses in North American swine and humans, 2009–2011. J. Virol 86, 8872–8878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson MI, Wentworth DE, Culhane MR, Vincent AL, Viboud C, LaPointe MP, Lin X, Holmes EC, Detmer SE, 2014. Introductions and evolution of human-origin seasonal influenza A viruses in multinational Swine populations. J. Virol 88, 10110–10119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson M, Culhane MR, Rovira A, Torremorell M, Guerrero P, Norambuena J, 2015a. Novel human-like influenza A viruses circulate in swine in Mexico and Chile. PLoS Curr. 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson MI, Schaefer R, Gava D, Cantao ME, Ciacci-Zanella JR, 2015b. Influenza A viruses of human origin in swine, Brazil. Emerg. Infect. Dis 21, 1339–1347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson MI, Viboud C, Vincent AL, Culhane MR, Detmer SE, Wentworth DE, Rambaut A, Suchard MA, Holmes EC, Lemey P, 2015c. Global migration of influenza A viruses in swine. Nat. Commun 6, 6696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ngo LT, Hiromoto Y, Pham VP, Le HT, Nguyen HT, Le VT, Takemae N, Saito T, 2012. Isolation of novel triple-reassortant swine H3N2 influenza viruses possessing the hemagglutinin and neuraminidase genes of a seasonal influenza virus in Vietnam in 2010. Influenza Other Respir. Viruses 6, 6–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pena L, Vincent AL, Ye J, Ciacci-Zanella JR, Angel M, Lorusso A, Gauger PC, Janke BH, Loving CL, Perez DR, 2011. Modifications in the polymerase genes of a swine-like triple-reassortant influenza virus to generate live attenuated vaccines against 2009 pandemic H1N1 viruses. J. Virol 85, 456–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereda A, Rimondi A, Cappuccio J, Sanguinetti R, Angel M, Ye J, Sutton T, Dibarbora M, Olivera V, Craig MI, Quiroga M, Machuca M, Ferrero A, Perfumo C, Perez DR, 2011. Evidence of reassortment of pandemic H1N1 influenza virus in swine in Argentina: are we facing the expansion of potential epicenters of influenza emergence? Influenza Other Respir. Viruses 5, 409–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petsch B, Schnee M, Vogel AB, Lange E, Hoffmann B, Voss D, Schlake T, Thess A, Kallen KJ, Stitz L, Kramps T, 2012. Protective efficacy of in vitro synthesized, specific mRNA vaccines against influenza A virus infection. Nat. Biotechnol 30, 1210–1216. [DOI] [PubMed] [Google Scholar]
- Quinnan GV, Schooley R, Dolin R, Ennis FA, Gross P, Gwaltney JM, 1983. Serologic responses and systemic reactions in adults after vaccination with monovalent A/USSR/77 and trivalent A/USSR/77, A/Texas/77, B/Hong Kong/72 influenza vaccines. Rev. Infect. Dis 5, 748–757. [DOI] [PubMed] [Google Scholar]
- Rajao DS, Loving CL, Gauger PC, Kitikoon P, Vincent AL, 2014. Influenza A virus hemagglutinin protein subunit vaccine elicits vaccine-associated enhanced respiratory disease in pigs. Vaccine 32, 5170–5176. [DOI] [PubMed] [Google Scholar]
- Rajao DS, Gauger PC, Anderson TK, Lewis NS, Abente EJ, Killian ML, Perez DR, Sutton TC, Zhang J, Vincent AL, 2015. Novel reassortant human-like H3N2 and H3N1 influenza A viruses detected in pigs are virulent and antigenically distinct from swine viruses endemic to the United States. J. Virol 89, 11213–11222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajao DS, Sandbulte MR, Gauger PC, Kitikoon P, Platt R, Roth JA, Perez DR, Loving CL, Vincent AL, 2016. Heterologous challenge in the presence of maternally-derived antibodies results in vaccine-associated enhanced respiratory disease in weaned piglets. Virology 491, 79–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richt JA, Lekcharoensuk P, Lager KM, Vincent AL, Loiacono CM, Janke BH, Wu WH, Yoon KJ, Webby RJ, Solorzano A, Garcia-Sastre A, 2006. Vaccination of pigs against swine influenza viruses using a NS1-truncated modified live virus vaccine. J. Virol 80, 11009–11018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell CA, Jones TC, Barr IG, Cox NJ, Garten RJ, Gregory V, Gust ID, Hampson AW, Hay AJ, Hurt AC, de Jong JC, Kelso A, Klimov AI, Kageyama T, Komadina N, Lapedes AS, Lin YP, Mosterin A, Obuchi M, Odagiri T, Osterhaus AD, Rimmelzwaan GF, Shaw MW, Skepner E, Stohr K, Tashiro M, Fouchier RA, Smith DJ, 2008. Influenza vaccine strain selection and recent studies on the global migration of seasonal influenza viruses. Vaccine 26 (Suppl. 4), D31–34. [DOI] [PubMed] [Google Scholar]
- Sandbulte MR, Spickler AR, Zaabel PK, Roth JA, 2015. Optimal use of vaccines for control of influenza A virus in swine. Vaccines (Basel) 3, 22–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon G, Larsen LE, Durrwald R, Foni E, Harder T, Van Reeth K, Markowska-Daniel I, Reid SM, Dan A, Maldonado J, Huovilainen A, Billinis C, Davidson I, Aguero M, Vila T, Herve S, Breum SO, Chiapponi C, Urbaniak K, Kyriakis CS, consortium E, Brown IH, Loeffen W, 2014. European surveillance network for influenza in pigs: surveillance programs, diagnostic tools and Swine influenza virus subtypes identified in 14 European countries from 2010 to 2013. PLoS One 9, e115815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith DJ, Lapedes AS, de Jong JC, Bestebroer TM, Rimmelzwaan GF, Osterhaus AD, Fouchier RA, 2004. Mapping the antigenic and genetic evolution of influenza virus. Science 305, 371–376. [DOI] [PubMed] [Google Scholar]
- Smith GJD, Vijaykrishna D, Bahl J, Lycett SJ, Worobey M, Pybus OG, Ma SK, Cheung CL, Raghwani J, Bhatt S, Peiris JSM, Guan Y, Rambaut A, 2009. Origins and evolutionary genomics of the 2009 swine-origin H1N1 influenza A epidemic. Nature 459, 1122–1125. [DOI] [PubMed] [Google Scholar]
- Solorzano A, Webby RJ, Lager KM, Janke BH, Garcia-Sastre A, Richt JA, 2005. Mutations in the NS1 protein of swine influenza virus impair anti-interferon activity and confer attenuation in pigs. J. Virol 79, 7535–7543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takemae N, Nguyen T, Ngo LT, Hiromoto Y, Uchida Y, Pham VP, Kageyama T, Kasuo S, Shimada S, Yamashita Y, Goto K, Kubo H, Le VT, Van Vo H, Do HT, Nguyen DH, Hayashi T, Matsuu A, Saito T, 2013. Antigenic variation of H1N1, H1N2 and H3N2 swine influenza viruses in Japan and Vietnam. Arch. Virol 158, 859–876. [DOI] [PubMed] [Google Scholar]
- Van Reeth K, Ma W, 2013. Swine influenza virus vaccines: to change or not to change-that’s the question. Curr. Top. Microbiol. Immunol 370, 173–200. [DOI] [PubMed] [Google Scholar]
- Van Reeth K, Brown IH, Olsen CW, 2012. Swine influenza. In: Zimmerman JJ (Ed.), Diseases of Swine. Wiley-Blackwell, Chichester West Sussex p.. [Google Scholar]
- Vander Veen RL, Loynachan AT, Mogler MA, Russell BJ, Harris DL, Kamrud KI, 2012. Safety, immunogenicity, and efficacy of an alphavirus replicon-based swine influenza virus hemagglutinin vaccine. Vaccine 30, 1944–1950. [DOI] [PubMed] [Google Scholar]
- Vander Veen RL, Mogler MA, Russell BJ, Loynachan AT, Harris DL, Kamrud KI, 2013. Haemagglutinin and nucleoprotein replicon particle vaccination of swine protects against the pandemic H1N1 2009 virus. Vet. Rec 173, 344. [DOI] [PubMed] [Google Scholar]
- Vijaykrishna D, Smith GJ, Pybus OG, Zhu H, Bhatt S, Poon LL, Riley S, Bahl J, Ma SK, Cheung CL, Perera RA, Chen H, Shortridge KF, Webby RJ, Webster RG, Guan Y, Peiris JS, 2011. Long-term evolution and transmission dynamics of swine influenza A virus. Nature 473, 519–522. [DOI] [PubMed] [Google Scholar]
- Vincent AL, Ma W, Lager KM, Janke BH, Webby RJ, Garcia-Sastre A, Richt JA, 2007. Efficacy of intranasal administration of a truncated NS1 modified live influenza virus vaccine in swine. Vaccine 25, 7999–8009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vincent AL, Ma W, Lager KM, Janke BH, Richt JA, 2008. Swine influenza viruses a North American perspective. Adv. Virus Res 72, 127–154. [DOI] [PubMed] [Google Scholar]
- Vincent AL, Ma W, Lager KM, Richt JA, Janke BH, Sandbulte MR, Gauger PC, Loving CL, Webby RJ, Garcia-Sastre A, 2012. Live attenuated influenza vaccine provides superior protection from heterologous infection in pigs with maternal antibodies without inducing vaccine-associated enhanced respiratory disease. J. Virol 86, 10597–10605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vincent A, Awada L, Brown I, Chen H, Claes F, Dauphin G, Donis R, Culhane M, Hamilton K, Lewis N, Mumford E, Nguyen T, Parchariyanon S, Pasick J, Pavade G, Pereda A, Peiris M, Saito T, Swenson S, Van Reeth K, Webby R, Wong F, Ciacci-Zanella J, 2014a. Review of influenza A virus in swine worldwide: a call for increased surveillance and research. Zoonoses Public Health 61, 4–17. [DOI] [PubMed] [Google Scholar]
- Vincent AL, Lager KM, Anderson TK, 2014b. A brief introduction to influenza a virus in Swine. Methods Mol. Biol 243–258. [DOI] [PubMed] [Google Scholar]
- Watson SJ, Langat P, Reid SM, Lam TT, Cotten M, Kelly M, Van Reeth K, Qiu Y, Simon G, Bonin E, Foni E, Chiapponi C, Larsen L, Hjulsager C, Markowska-Daniel I, Urbaniak K, Durrwald R, Schlegel M, Huovilainen A, Davidson I, Dan A, Loeffen W, Edwards S, Bublot M, Vila T, Maldonado J, Valls L, Consortium E, Brown IH, Pybus OG, Kellam P, 2015. Molecular epidemiology and evolution of influenza viruses circulating within European swine between 2009 and 2013. J. Virol 89, 9920–9931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wesley RD, Lager KM, 2006. Overcoming maternal antibody interference by vaccination with human adenovirus 5 recombinant viruses expressing the hemagglutinin and the nucleoprotein of swine influenza virus. Vet. Microbiol 118, 67–75. [DOI] [PubMed] [Google Scholar]
- Wiersma LC, Rimmelzwaan GF, de Vries RD, 2015. Developing universal influenza vaccines: hitting the nail, not just on the head. Vaccines (Basel) 3, 239–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiley DC, Wilson IA, Skehel JJ, 1981. Structural identification of the antibody-binding sites of Hong Kong influenza haemagglutinin and their involvement in antigenic variation. Nature 289, 373–378. [DOI] [PubMed] [Google Scholar]
- Worobey M, Han GZ, Rambaut A, 2014. Genesis and pathogenesis of the 1918 pandemic H1N1 influenza A virus. Proc. Natl. Acad. Sci. U. S. A 111, 8107–8112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- York I, Donis RO, 2013. The 2009 pandemic influenza virus: where did it come from, where is it now, and where is it going? Curr. Top. Microbiol. Immunol 370, 241–257. [DOI] [PubMed] [Google Scholar]
- Zell R, Scholtissek C, Ludwig S, 2013. Genetics, evolution, and the zoonotic capacity of European Swine influenza viruses. Curr. Top. Microbiol. Immunol 370, 29–55. [DOI] [PubMed] [Google Scholar]
- Zhu H, Webby R, Lam TT, Smith DK, Peiris JS, Guan Y, 2013. History of Swine influenza viruses in Asia. Curr. Top. Microbiol. Immunol 370, 57–68. [DOI] [PubMed] [Google Scholar]


