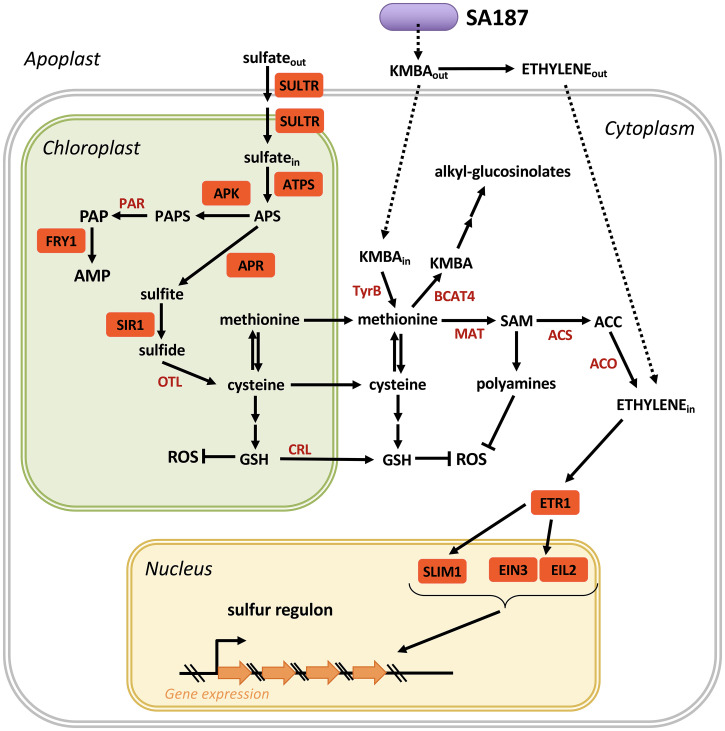
Fig. 5.
Model of SA187-induced sulfur metabolism and ethylene signaling in Arabidopsis: uptake of sulfate is followed by conversion into APS, sulfite, sulfide, and cysteine, which can be converted into either glutathione (GSH) or methionine. GSH can block salt-induced ROS production in plastids. Methionine can be converted into SAM, ACC, and ethylene, which regulates the sulfur regulon via the transcription factors SLIM1, EIL2, and EIN3. The ethylene-regulated sulfur regulon comprises most genes of the sulfur metabolism, thereby creating a feedback loop between sulfur status and genes encoding sulfur metabolic enzymes. Sulfur status is conveyed by PAP level–induced retrograde signaling to regulate nuclear encoded plastid and sulfur metabolic genes. SULTR = sulfate transporter, ATPS = ATP sulfurylase, APR = APS reductase, APK = APS kinase, PAR = PAPS reductase, SIR1 = sulfite reductase, FRY1 = 5′-bisphosphate, nucleotide phosphatase, OTL = O-acetylserine thiollyase, TyrB = aromatic–amino acid transaminase, BCAT4 = methionine transaminase, MAT = methionine adenosyltransferase, ACS = ACC synthase, ACO = ACC oxidase, ETR1 = ethylene receptor 1, SLIM1 = sulfur limitation1 (central transcriptional regulator of plant sulfur response and metabolism), EIN3 = ethylene-insensitive 3, EIL2 = ethylene-insensitive 3-like 2, CRL = chloroquine-resistance transporter (CRT)-like protein, APS = adenosine 5′-phophosulfate, PAPS = 3′-phosphoadenosine-5′-phosphosulfate, PAP = 3′-polyadenosine 5′-phosphate, AMP = adenosine 5′-monophophfate, KMBA = 2-keto-4-methylthiobutyric acid, SAM = S-adenosyl methionine, ACC = 1-aminocyclopropane-1-carboxylic acid, GSH = glutathione, ROS = reactive oxygen species.