Fig. 2.
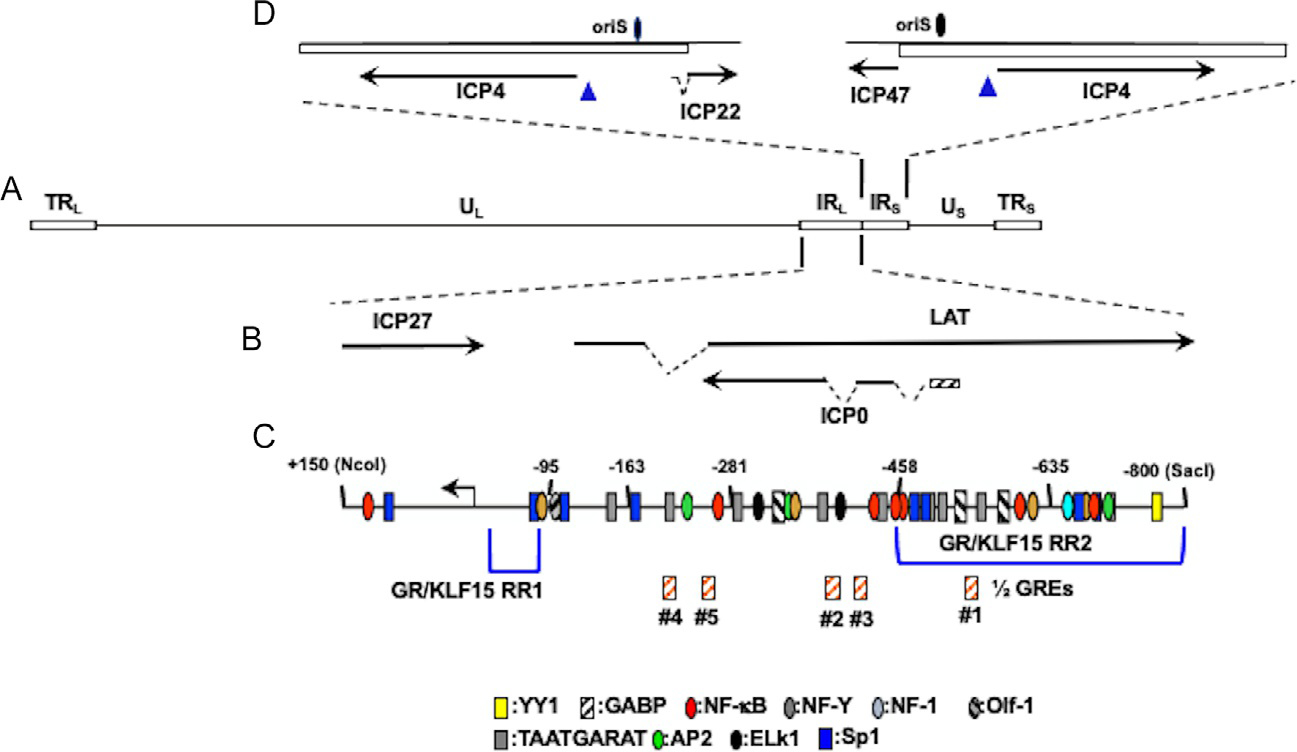
Location of ICP4 gene, ICP0 gene, and adjacent genes within HSV-1 genome. Panel (A) The prototypic HSV-1 genomic structure is shown. Viral repeat regions are shown as open rectangles. TRL is the terminal long repeat. IRL is the internal (or inverted) long repeat. TRS is the terminal short repeat. IRS is the internal (or inverted) short repeat. The unique long (UL) and unique short (US) regions are depicted by a solid line. Panel (B) Location of ICP0 and other genes within IRL. Gene organization within TRL is the same as IRL. Dashed lines denote introns in ICP22, LAT, and ICP0. Panel (C) Schematic of ICP0 promoter (−800 to +150). Numbers assigned correspond to nucleotide positions relative to the transcription initiation site (black arrow). Location of ½ GREs is indicated by orange hatched boxes. GR and KLF15 responsive regions (GR/KLF15 RR1 and GR/KLF15 RR2) previously identified in Neuro-2A and Vero cells are denoted by blue brackets (Ostler et al., 2019; Sinani, Cordes, Workman, Thunuguntia, & Jones, 2013). Location of other potential transcription factor binding sites were previously identified (Kushnir, Davido, & Schaffer, 2009). Panel (D) Location of ICP4 coding regions and flanking genes in IRS (ICP22) or TRS (ICP47). Dashed lines in ICP22 denote the location of an intron. Location of the origin of replication oriS is denoted by black oval.
