Fig. 1 |. Overview of the workflow for mitochondrial ribosome profiling.
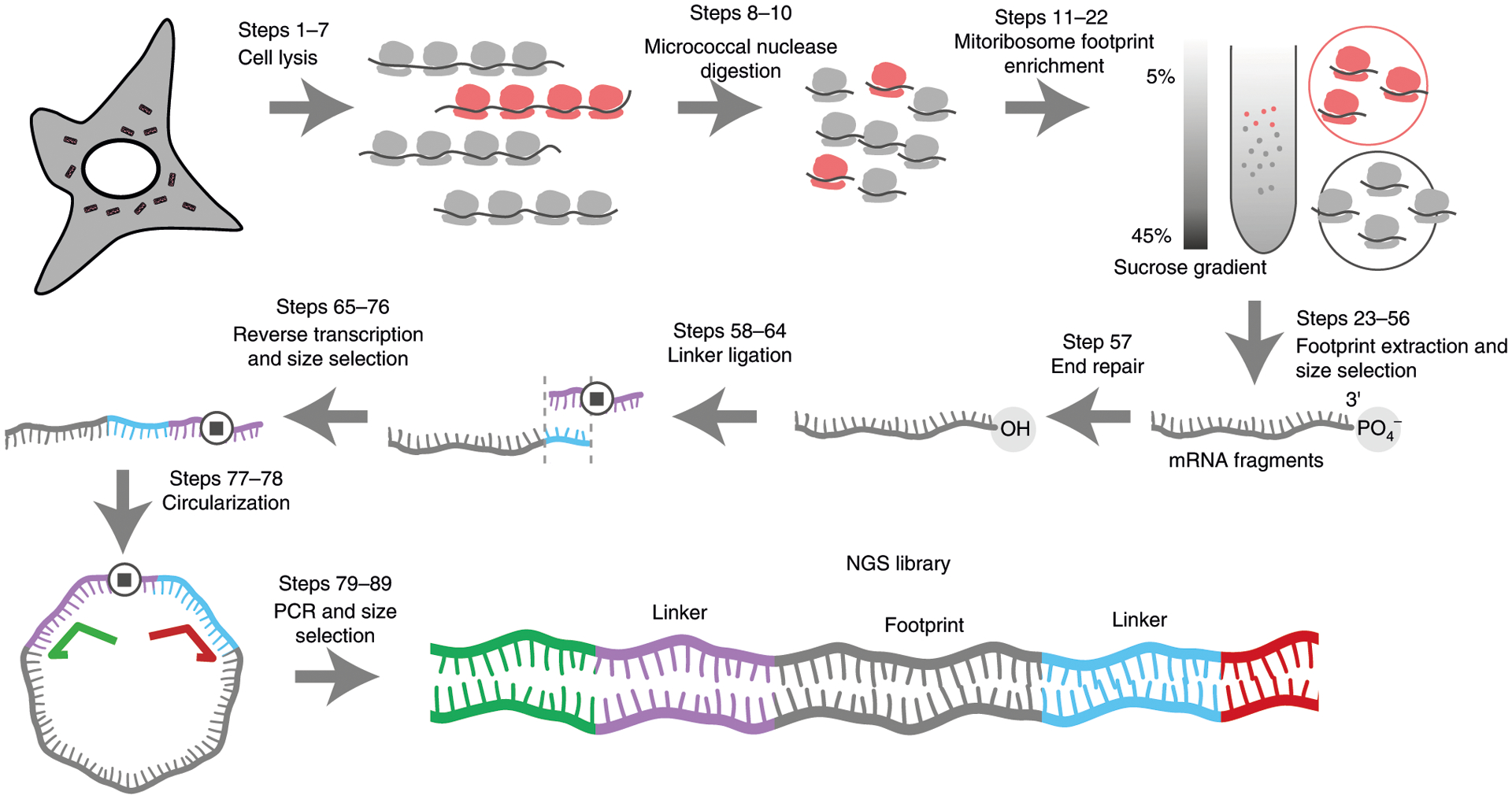
Steps 1–3: mitoribosome enrichment. Cell lysates contain both cytosolic (represented in gray) and mitochondrial (represented in red) ribosomes. After nuclease digestion, the cell lysate is layered onto a 5–45% sucrose gradient and ultracentrifuged. Cytosolic and mitochondrial ribosomes have different sedimentation coefficients and can therefore be separated by ultracentrifugation and collected separately. Steps 4–9: library preparation after the mRNA fragments are extracted. The 3′-end is first repaired to remove the 3′-end phosphate group followed by a 3′-linker ligation (linker in blue). A reverse transcription primer (purple) with a complementary sequence to the linker is used to generate cDNA. The stop symbol represents a spacer to avoid circular amplification during the PCR step. The ssDNA is circularized and amplified to generate NGS library using a universal forward primer (green) and a reverse primer (red) with Illumina barcodes.
