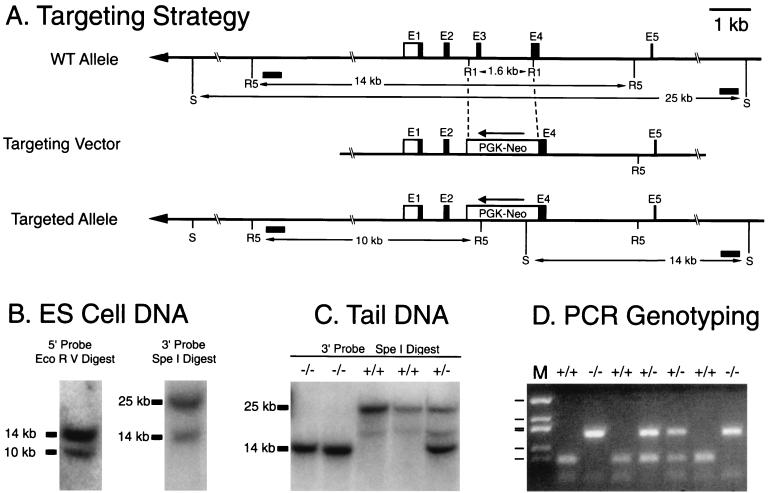
FIG. 1.
mPer3 targeting construct and genotyping strategies. (A) Schematic representation of the mPer3 gene, the targeting construct, and the targeted allele. A 1.6-kb portion of the mPer3 gene was excised with EcoRI (R1), and a PGK-NEO cassette was inserted in the reverse orientation. The 5′ and 3′ arms of the targeting construct were ca. 6 and 9 kb, respectively. Restriction sites introduced by the PGK-NEO cassette and used for Southern blot analysis were EcoRV (R5) and SpeI (S). Intron and exon structure is shown only for the first five exons (E1 to E5). Boxes represent exons, and the lines between the boxes indicate introns. Open boxes represent untranslated regions. Filled boxes represent putative coding regions based on translation from the first ATG with a Kozak consensus sequence at nt 358 of AF050182. (A second Kozak consensus sequence present in exon 2 could begin translation at nt 523). Horizontal filled boxes indicate the locations of probes used for Southern blotting. (B) Southern blot identification of the targeting event in ES cells. Representative autoradiograms are shown. The upper band in each case represents the wild-type allele, and the lower band represents the targeted allele (see panel A). (C) Southern blot analysis of DNA extracted from mouse tails. Genotypes are shown above lanes. (D) PCR genotyping of DNA extracted from mouse tails. Genotypes are shown above lanes. M, size marker (pUC19 DdeI with bands at 910, 540, 426, 409, 266, and 166 bp).