Figure 2.
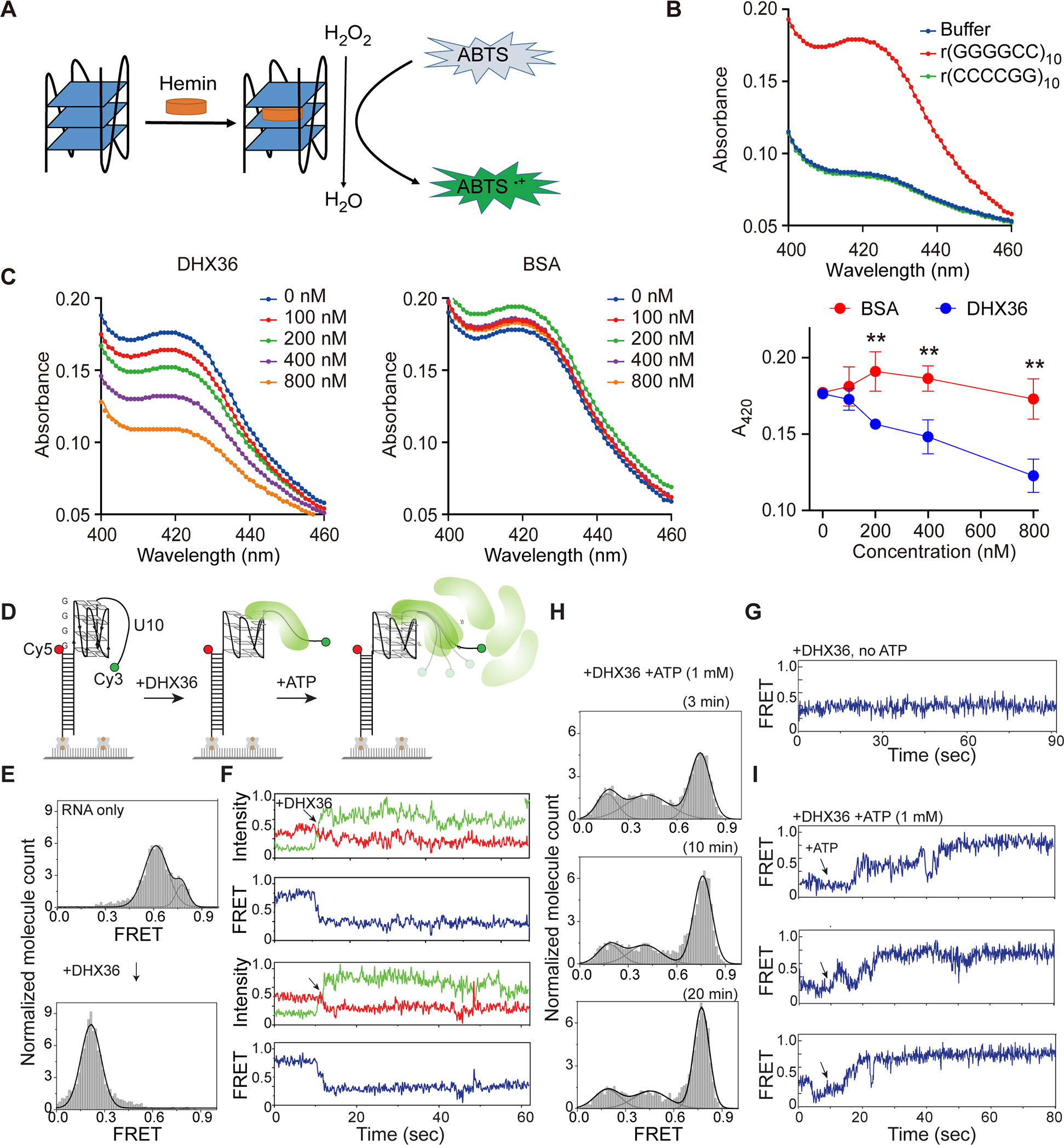
DHX36 efficiently unwinds G-quadruplexes formed by GGGGCC RNA repeats. (A) Schematic representation of the G-quadruplex-hemin RNAzyme system. (B) Absorption spectra of r(GGGGCC)10, r(CCCCGG)10, or the buffer control in the colorimetric reactions. [RNA]=1 μM, [Hemin]=2.5 μM, [ABTS]=2.5 μM, and [H2O2]=0.4 mM. (C) DHX36 or BSA proteins were added to the r(GGGGCC)10 RNAzyme system at increasing concentrations (0, 100, 200, 400, or 800 nM) before triggering the colorimetric reaction. Maximum absorbance at 420 nm for each group was used to compare the RNAzyme activities. Data are given as means ± SD of four independent experiments. **P <0.01. (D) Schematic of smFRET using RNA r(GGGGCC)4-U10. (E) FRET histograms before and after addition of the DHX36 protein. FRET values were collected from >4000 molecules in 20 different fields of view. (F) Two sets of representative smFRET traces upon addition of DHX36 (arrows). The smFRET trace shows fluorescence intensities observed for Cy3 (green) and Cy5 (red) and the calculated FRET efficiency (blue) when 10 nM DHX36 protein was added. (G) The smFRET trace taken after DHX36 was added to the G4C2-U10 substrate. (H) FRET histograms taken 3, 10, and 20 minutes after 1 mM ATP was added to the DHX36 and G4C2-U10 substrate mix. (I) Three sets of representative smFRET traces after ATP was added.
