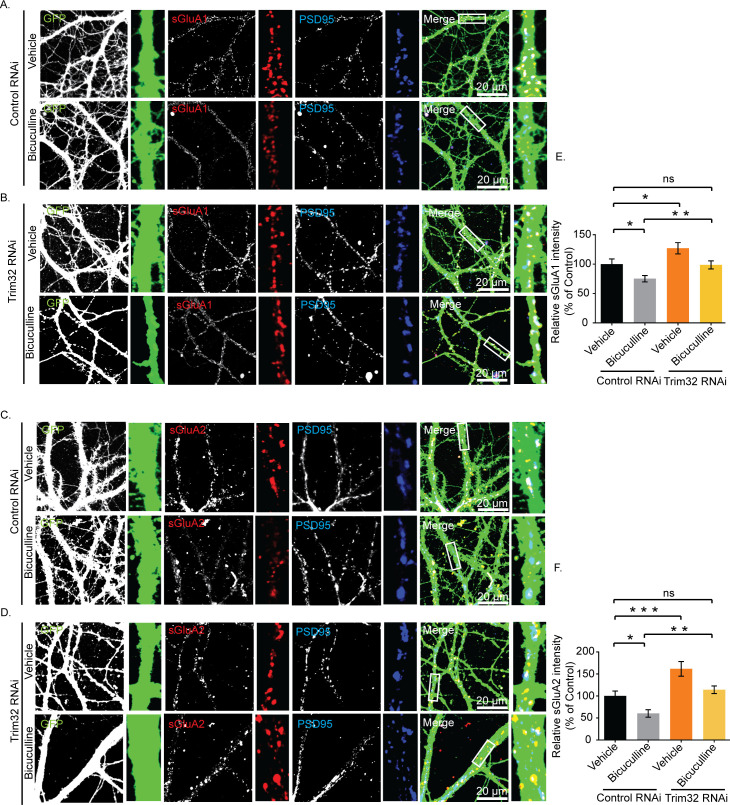
Fig 12. Trim32-mediated synaptic downscaling occurs via modulation of sAMPARs.
Neurons were transduced with lentivirus coexpressing EGFP and shRNA against Trim32 or a nontargeting shRNA and the expression of sGluA1 and sGluA2 determined by immunostaining. (A, B) High-magnification images of bicuculline- and vehicle-treated neurons expressing nontargeting shRNA (Control RNAi) (A) or expressing shRNA against Trim32 (Trim32 RNAi) (B) showing the expression of sGluA1 (red), PSD95 (blue), GFP (green), and GFP/sGluA1/PSD95 (merged). (C, D) High-magnification images of bicuculline- and vehicle-treated neurons expressing nontargeting shRNA (Control RNAi) (C) or shRNA against Trim32 (Trim32 RNAi) (D) showing the expression of sGluA2 (red), PSD95 (blue), GFP (green), and GFP/sGluA2/PSD95 (merged). (E) Quantitation of normalized intensity of synaptic sGluA1. (F) Quantitation of normalized intensity of synaptic sGluA2. n = 30–40 for GluA1; n = 23–36 for GluA2. Data shown as mean ± SEM. *p < 0.02, **p < 0.03, for sGluA1. *p < 0.008, **p < 0.002, ***p < 0.0004 for sGluA2. One-way ANOVA and Fisher’s LSD. Dendrite marked in white box was digitally amplified. See also S5 Fig. The data underlying this figure are available at https://figshare.com/articles/dataset/Homeostatic_scaling_is_driven_by_a_translation-dependent_degradation_axis_that_recruits_miRISC_remodeling/16768816. ns, not significant; sAMPAR, surface AMPAR.