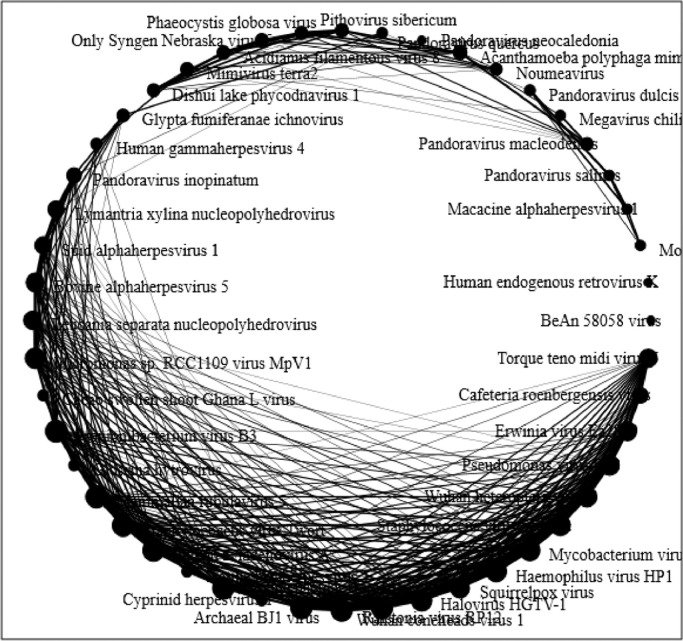
Fig. 9.
Network Analysis: Using average distance matrix based on the tetranucleotide frequencies a networking circular model for the metaviromes was calculated. Each viral metagenomic reads (edge cut off at 80%) are represented as singular nodal points and the node size is governed by the weight of the edges connecting the node. The edges of the network shows the correlation between the viral genomes and their nucleotide frequency abundances. The weight of the edges correlates the frequency between the nodes.