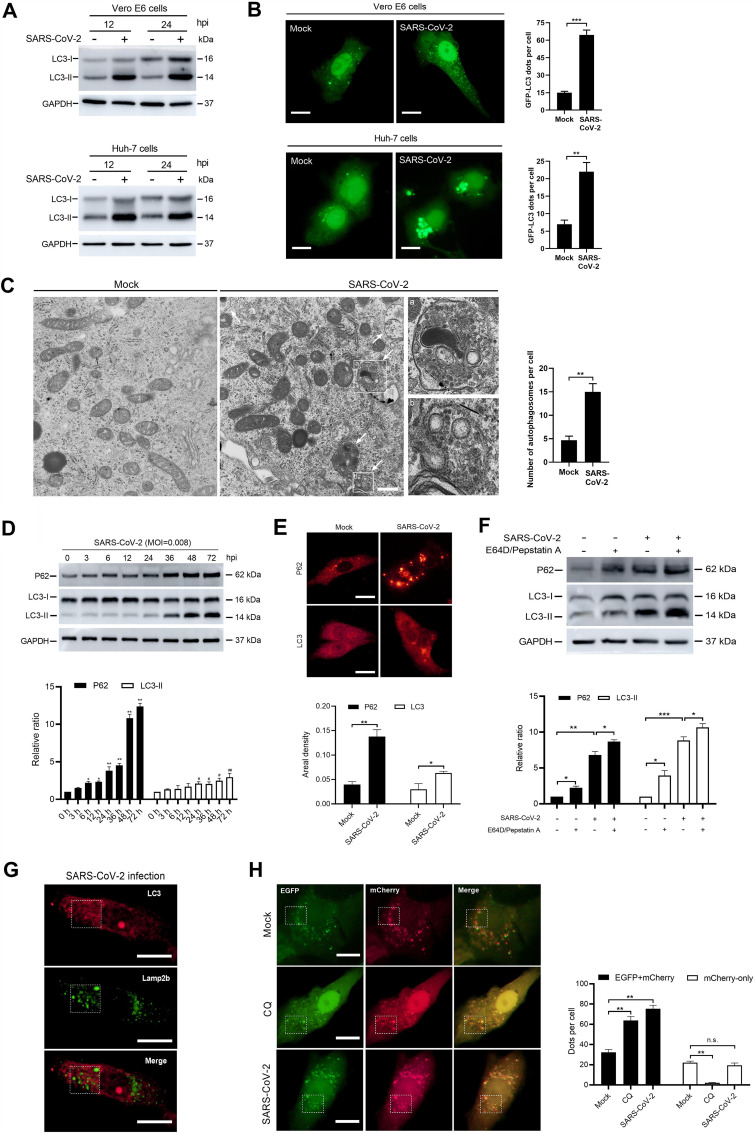
FIG 2.
SARS-CoV-2 increases autophagosome formation but blocks autophagosome-lysosome fusion. Vero E6 and Huh-7 cells were treated with a vehicle or SARS-CoV-2 at an MOI of 0.008. (A) LC3 expression at 12 and 24 hpi in Vero E6 and Huh-7 cells. Samples were analyzed by Western blotting. (B) GFP-LC3 accumulation in mock- and SARS-CoV-2-infected Vero E6 and Huh-7 cells at 24 hpi. Cells were transfected with GFP-LC3 plasmids and analyzed for autophagosome accumulation. Theamount of GFP-LC3 per cell was calculated from at least 30 cells from each group. Scale bar = 5 μm. (C) Electron microscope images of autophagosomes (white arrows) in Vero E6 cells at 6 hpi. The number of autophagosomes per cell was calculated from at least 10 cells. Scale bar = 500 nm. (D) p62 and LC3 expression in Vero E6 cells at indicated time points after SARS-CoV-2 infection. Samples were analyzed by Western blotting. (E) Immunofluorescence analysis of p62 and LC3 in mock- or SARS-CoV-2-infected Vero E6 cells at 24 hpi. The areal density of p62 and LC3 was calculated from at least 30 cells for each group. Scale bar = 5 μm. (F) Western blotting of p62 and LC3 expression in Vero E6 cells that had been treated or not for 2 h with E64D (10 μg/ml) and pepstatin A (10 μg/ml) before mock or SARS-CoV-2 infection. Samples were collected at 24 hpi. (G) Immunofluorescence analysis of LC3 and lamp2b colocalization in SARS-CoV-2-infected Vero E6 cells at 24 hpi. Scale bar = 5 μm. (H) Colocalization analysis of EGFP and mCherry in mock-infected, CQ-treated, and SARS-CoV-2-infected Vero E6 cells, pretransfected with EGFP-mCherry-LC3 plasmids. In the CQ group, 40 μM drug was added to the cells 2 h before infection. Samples were collected at 24 hpi. The numbers of EFGP-mCherry (yellow) and mCherry-only dots were calculated from at least 30 cells from each group. Scale bar = 5 μm. Data were expressed as means ± SEM from three experiments. *, P < 0.05; **, P < 0.01; ***, P < 0.001. n.s., not significant.