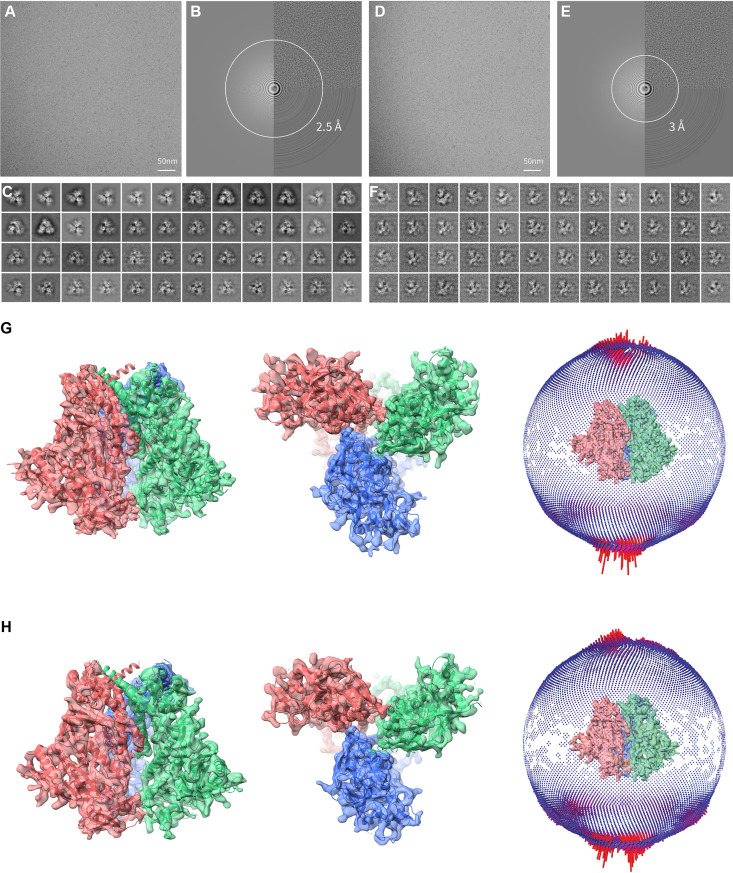
FIG 4.
Cryo-EM analysis of the full-length HIV-1JR-FL Env(−) trimer. (A) A typical cryo-EM micrograph of Env(−) trimers taken with a Gatan K2 direct electron detector at 0 degrees of tilt. (B) Fourier transform of the image in panel A. Left panel, simulated logarithmic amplitude spectra in Gctf (134); upper right panel, background-subtracted logarithmic amplitude spectra; lower right panel, equiphase average in Gctf. (C) Unsupervised 2D class averages for nontilt particles. (D) A typical cryo-EM micrograph of Env(−) trimers taken with a Gatan K2 direct electron detector at 45 degrees of tilt. (E) Fourier transform of the image in panel D. Left panel, simulated logarithmic amplitude spectra in Gctf (134); upper right panel, background-subtracted logarithmic amplitude spectra; lower right panel, equiphase average in Gctf. (F) Unsupervised 2D class averages for tilted particles. (G) Final refined cryo-EM density map for the state-U1 Env(−) trimers. Left, side view, with gp120 at the bottom of the figure and gp41 at the top. Middle, top view from the gp120 side. The right inset shows the orientation distribution of the particles used for reconstruction of the final state-U1 map. (H) Final refined cryo-EM density map for the state-U2 Env(−) trimers. Left, side view, with gp120 at the bottom of the figure and gp41 at the top. Middle, top view from the gp120 side. The right inset shows the orientation distribution of the particles used for reconstruction of the final state-U2 map.