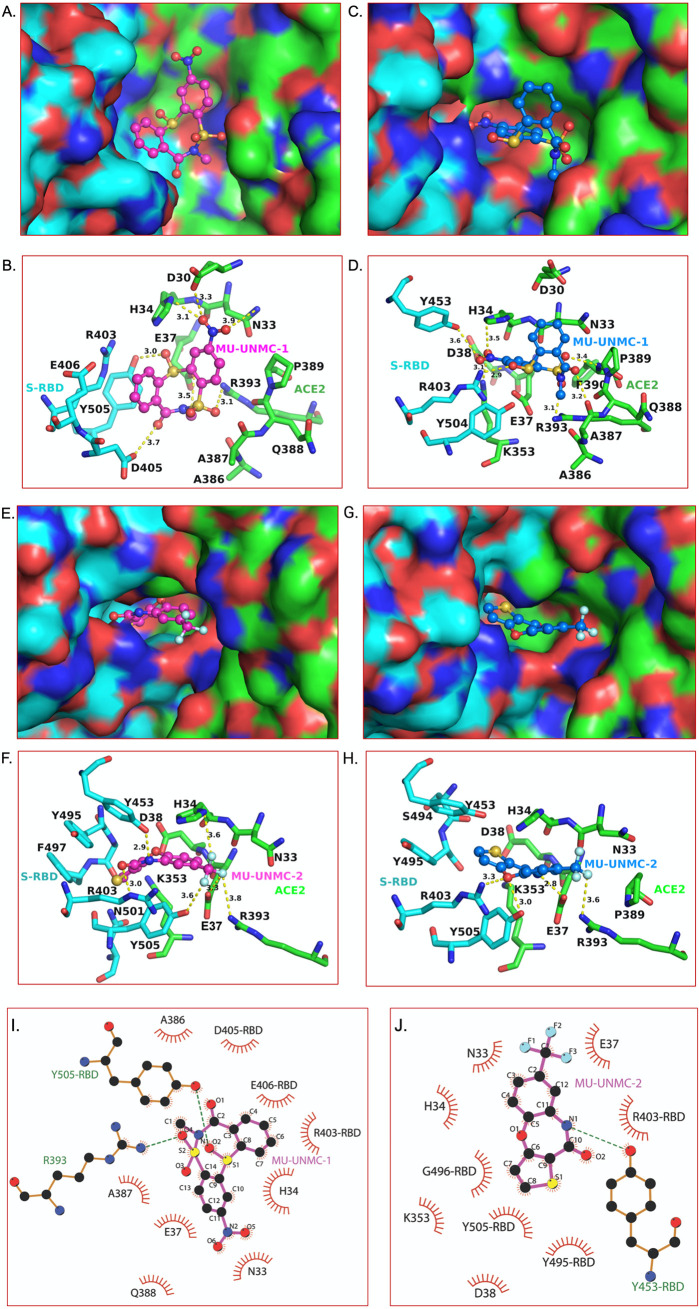
FIG 8.
Optimal binding mode of MU-UNMC-1 and MU-UNMC-2 with S-RBD/ACE2 complexes of Wuhan-Hu-1 and B.1.351. The best docking pose of MU-UNMC-1 and MU-UNMC-2 was obtained from the IFD. Panels A and C show surface representation of Wuhan-Hu-1 and B.1.351 S-RBD (cyan)/ACE2 (green) complexes, respectively for MU-UNMC-1. (B, D) The details of interactions of MU-UNMC-1 with S-RBD/ACE2 complexes of Wuhan-Hu-1 and B.1.351 are shown in panels B and D. Panels E and G show surface representation Wuhan-Hu-1 and B.1.351 S-RBD (cyan)/ACE2 (green) complexes, respectively for MU-UNMC-2. (F, H) The details of interactions of MU-UNMC-2 with S-RBD/ACE2 complexes of Wuhan-Hu-1 and B.1.351 are shown in panels F and H. The yellow dotted lines represent the polar interactions between MU-UNMC-2 and proteins. All colors and the representations are the same as in Fig. 1, except the teal carbons of MU-UNMC-1/2 in the S-RBD/ACE2 complex of the B.1.351 variant. The two panels represent the different modes of binding of MU-UNMC-1 and MU-UNMC-2 in the two variants. (Panels I and J show the hydrophobic and Van der Waals interactions of MU-UNMC-1 and MU-UNMC-2, respectively, with the Wuhan S-RBD/ACE2 complex. The S-RBD residues are distinguished from ACE2 by marking RBD after the residue number. R393 of ACE2 and Y505 of S-RBD also form the H-bond with MU-UNMC-2, and green dotted lines represent them. Similarly, Y453 of S-RBD forms an H-bond with the N1 atom MU-UNMC-2 and is shown as a green dotted line. The circle size is proportional to the interacting atoms from each residue and the atoms that interact are represented by ‘rays’ emerging from the atoms.