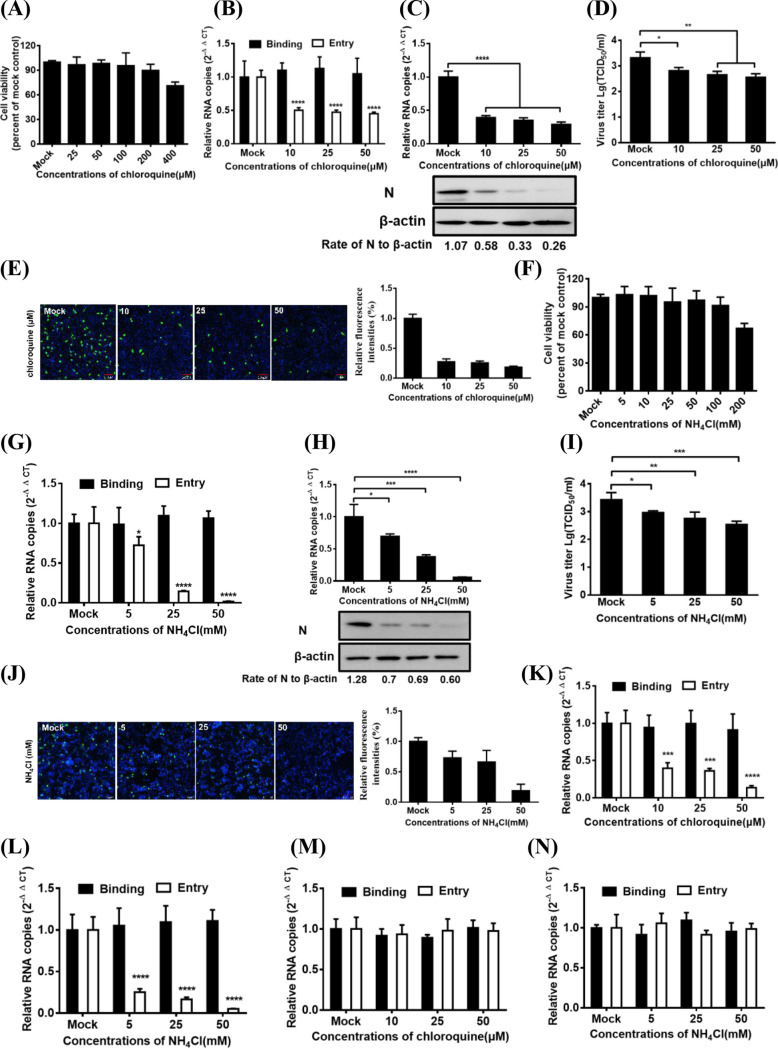
FIG 1.
PDCoV entry into IPI-2I cells requires an acidic endosomal pH. (A) CCK-8-based cell viability assay for chloroquine as described in Materials and Methods. (B) Chloroquine inhibited PDCoV entry but not binding. IPI-2I cells were pretreated with subtoxic doses at 37°C for 1 h and infected with PDCoV (MOI of 5) at 4°C for 1 h (binding step) and then shifted to 37°C for 1 h (entry step). Cells were lysed to determine viral RNA copy numbers by RT-qPCR. (C and D) RT-qPCR, Western blotting, and viral titer detection analysis for viral infection inhibition by chloroquine. Cells were pretreated with increasing subtoxic doses of chloroquine or DMSO at 37°C for 1 h and then inoculated with PDCoV (MOI of 5) at 37°C for 6 h. Cells were lysed to determine viral RNA copy numbers via RT-qPCR, N protein levels via Western blotting (C), and viral titer (D) via TCID50 assay. (E) IFA for viral infection inhibition by chloroquine. Cells were pretreated with subtoxic doses of these drugs at 37°C for 1 h and then inoculated with PDCoV (MOI of 5) at 37°C for 6 h. Cells were fixed and subjected to IFA. Relative fluorescence intensity was quantified by Image‐Pro Plus software as shown on the right. (F) CCK-8-based cell viability assay for NH4Cl as described in Materials and Methods. (G to J) RT-qPCR, Western blot analysis, viral titer detection, and IFA for viral entry and infection inhibition by NH4Cl. Cells were pretreated with increasing subtoxic doses of NH4Cl or H2O, followed by RT-qPCR (G), Western blotting (H), viral titer detection (I), and IFA (J) as described in corresponding panels B, C, D, and E. (K to N) Effect of chloroquine and NH4Cl on the entry of VSV-GFP and SeV into IPI-2I cells. The cells were pretreated with increasing subtoxic doses of chloroquine (K and M) or NH4Cl (L and N) and then were infected with VSV-GFP (K and L) and SeV (M and N) at 4°C for 1 h (binding step) and then shifted to 37°C for 1 h (entry step). Cells were lysed to determine viral RNA copy numbers by RT-qPCR. Target protein expression was quantitatively estimated by ImageJ software and presented as the density value relative to that of the β-actin. The presented results represent the means and standard deviations of data from three independent experiments. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001.