Figure 1.
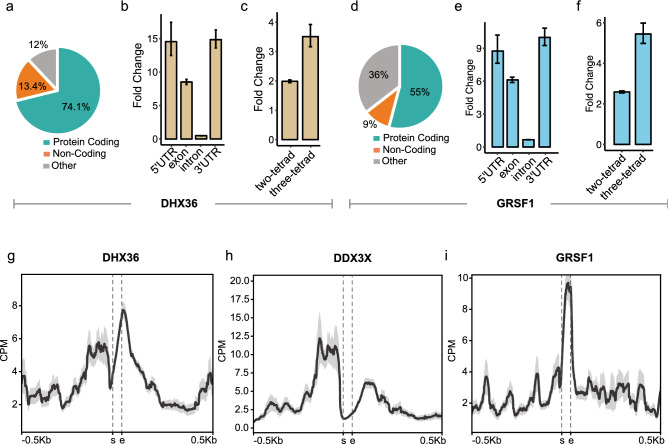
RNA binding proteins interact with G4s in mRNA. (a,d) Incidence of iCLAE peaks within protein-coding, non-coding or unannotated regions (other) of the genome. (b,e) Enrichment of iCLAE peaks at annotated RNA features. (c,f) Enrichment of iCLAE peaks at predicted G4 motifs. Enrichment calculated following random shuffling of peaks in the transcriptome (FDR = 1 × 10−4). Error bars represent 95% confidence interval. (g–i) Distribution of DHX36, DDX3X and GRSF1 iCLAE reads relative to G4 motifs. Graphs denote pileups of iCLAE reads from multiple sites in CPM. DDX3X iCLAE data has been previously published11.