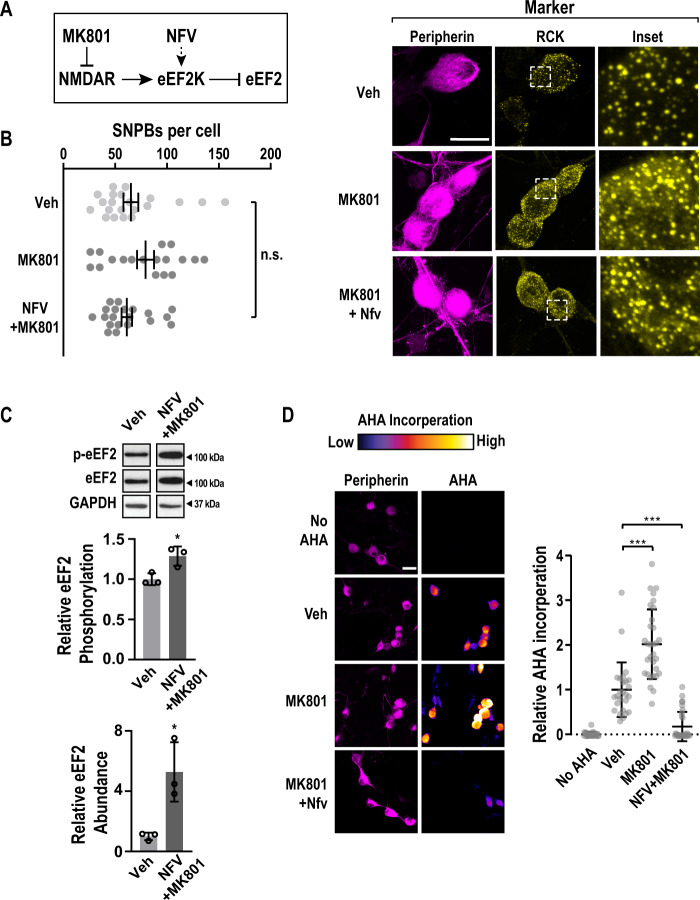
Fig. 4. Inhibition of NMDA receptors counteracts nelfinavir-induced PB loss.
A A schematic that indicates the relationship between MK801, an NMDAR antagonist, Nelfinavir (NFV), eEF2K, and eEF2. B left Primary DRG cultures were treated with vehicle (Veh), MK801 (10 µM), or co-treated with nelfinavir and MK801 (NFV+MK801) for a period of 1 h, and subjected to ICC. Confocal imaging was used to identify p-bodies and key markers. DRG neurons were identified by peripherin immunofluorescence (magenta) and SNPBs were identified based on Rck (yellow). Nuclei were stained with DAPI (cyan). B left Representative confocal images. Scale bar = 20 µm. B right Quantification of PBs per peripherin-positive cell. For Veh, MK801, and MK801 + NFV, n = 21, 18, and 20 cells, respectively. Bars indicate mean ± SEM. P-values determined by one-way ANOVA. C Primary DRG cultures were treated with vehicle (Veh) or co-treated with nelfinavir and MK801 (NFV+MK801). Lysates from treated cells were probed for p-eEF2, eEF2, and GAPDH (load control). C upper Representative immunoblots (cropped to depict one representative band per condition). C middle Quantification of mean p-eEF2/eEF2 signal. C lower Quantification of mean eEF2 normalized to GAPDH, n = 3 biological replicates. Error bars represent ±SD. P-values determined by Unpaired two-tailed t test. Upper panel p = 0.329, lower panel p = 0.0204. D Primary DRG cultures were subjected wo the same treatments as in B with the addition of a 30-min pulse of AHA. Cultures were then used for FUNCAT as well as peripherin immune-labeling before confocal imaging. As a control, a no-AHA group was also imaged. D left Representative confocal images. Scale bar = 30 µm. D right Quantification of AHA incorporation in peripherin positive cells. For No AHA, Veh, MK801, and MK801 + NFV, n = 29, 26, 31, and 31 cells, respectively. Bars indicate mean ±SD. P-values determined by one-way ANOVA. Veh vs. MK801 p < 0.0001, Veh vs. MK801 + NFV p < 0.0001. Source data are provided as a Source Data file. Uncropped blots are presented in Supplemental Fig. 7.