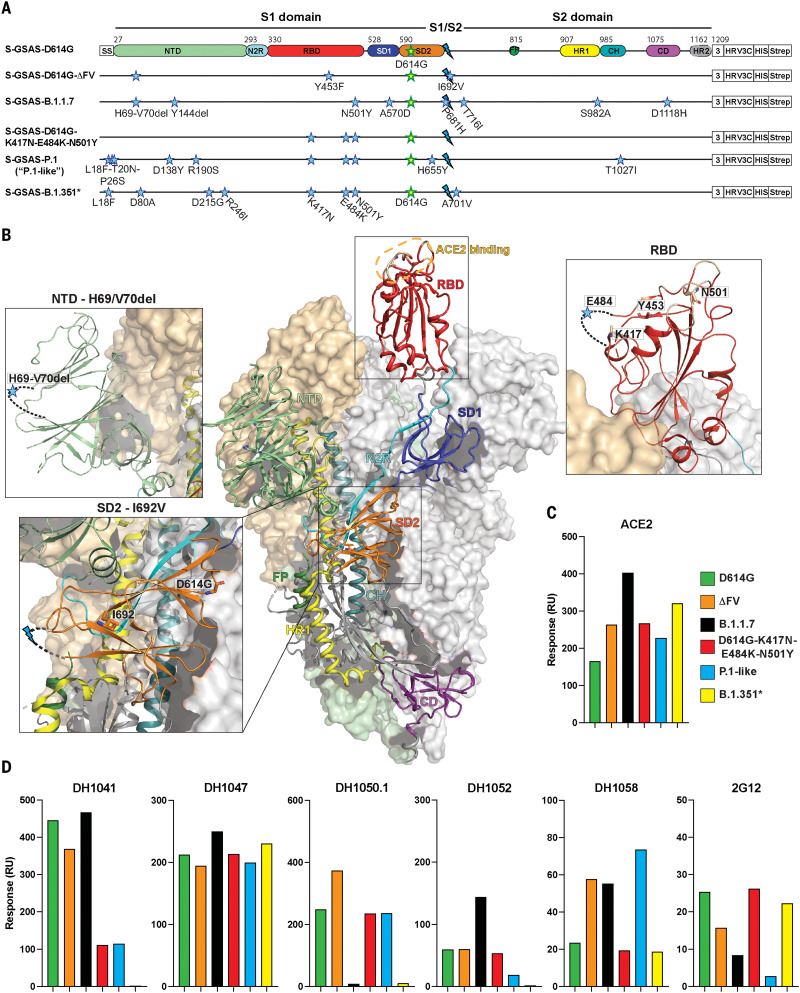
Fig. 1. SARS-CoV-2 spike (S) protein ectodomains for characterizing structures and antigenicity of S protein variants.
(A) Domain architecture of the SARS-CoV-2 spike protomer. The S1 subunit contains a signal sequence (SS), the NTD (N-terminal domain, pale green), N2R (NTD-to-RBD linker, cyan), RBD (receptor binding domain, red), and SD1 and SD2 (subdomains 1 and 2, dark blue and orange). The S2 subunit contains the FP (fusion peptide, dark green), HR1 (heptad repeat 1, yellow), CH (central helix, teal), CD (connector domain, purple), and HR2 (heptad repeat 2, gray) regions. The transmembrane domain (TM) and cytoplasmic tail (CT) have been truncated and replaced by a foldon trimerization sequence (3), an HRV3C cleavage site (HRV3C), a His-tag (HIS), and a strep-tag (Strep). The D614G mutation (yellow star with green outline) is in SD2. The S1/S2 furin cleavage site (RRAR) has been mutated to GSAS (blue lightning). The substitutions in each variant are indicated by blue stars. *A few ectodomain constructs were prepared on the B.1.351 spike backbone; these differed in their NTD mutations (see table S1). Binding data for the other constructs, including the one representing the dominant circulating form (L18F, D80A, D215G, Δ242-244, K417N, E484K, N501Y, D614G, A701V), are shown in figs. S2 and S3. The construct shown here was used for determining the cryo-EM structure (Fig. 6). The “P.1-like” spike was prepared in the P.1 backbone but retained the K417N RBD substitution (instead of the K417T in the P.1 spike; see table S1). (B) Representation of the trimeric SARS-CoV-2 spike ectodomain in a prefusion conformation with one RBD up (PDB ID 7KDL). The S1 subunit on an RBD-down protomer is shown as a pale orange molecular surface; the S2 subunit is shown in pale green. The subdomains on an RBD-up protomer are colored according to (A) on a ribbon diagram. Each inset corresponds to the spike regions harboring mutations included in this study. (C and D) Binding of ACE2 (C) and of RBD-directed antibodies DH1041 and DH1047, NTD-directed antibodies DH1050.1 and DH1052, and S2-directed antibodies DH1058 and 2G12 (D) to spike variants measured by SPR. Data are representative of two independent experiments.