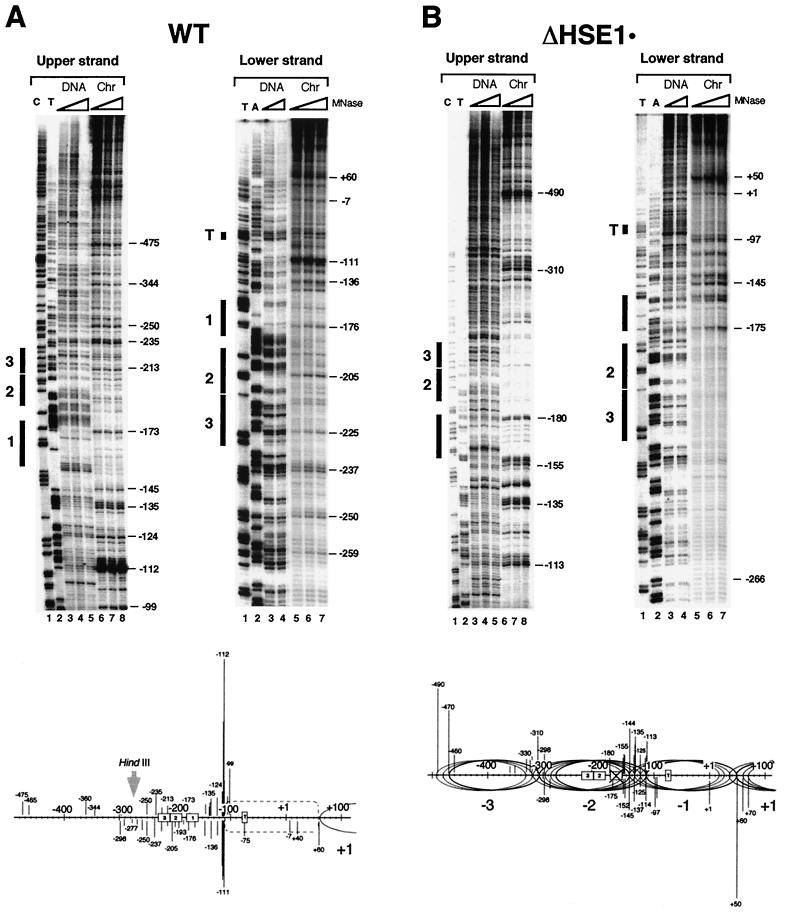
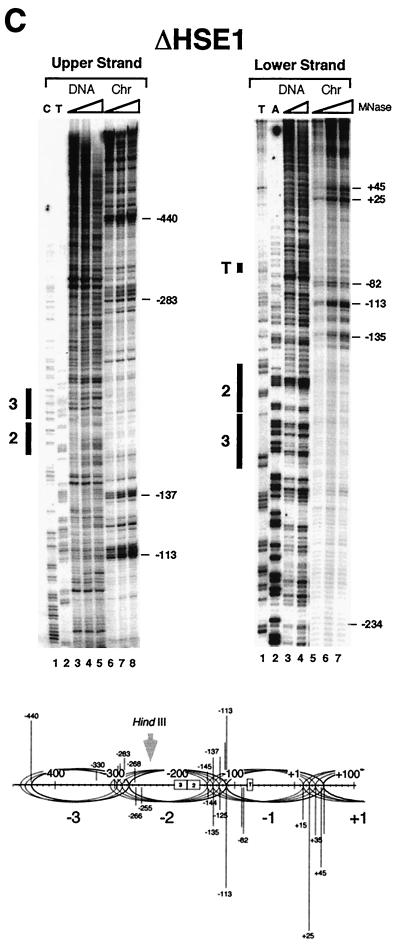
FIG. 2.
MNase genomic footprinting reveals that the promoter region of hsp82 alleles lacking HSE1 is assembled into a novel dinucleosome. Illustrated are strand-specific genomic footprints for HSP82+, hsp82-ΔHSE1 · (a 32-bp substitution mutant), and hsp82-ΔHSE1 (a 32-bp deletion mutant) (A to C, Chr lanes). Cells were cultivated at 30°C, nuclei were purified and digested with MNase, and genomic DNA was isolated. For the naked DNA controls (DNA), genomic DNA was isolated from each strain and digested with MNase. Each digestion series represents a set of twofold serial dilutions. Cut sites were mapped by linear PCR using both upper-strand- and lower-strand-specific primers (+26→−11 and −342→−315, respectively). Cleavage profiles were visualized using a PhosphorImager, and intensities of all major cut sites were quantitated. They are schematically illustrated below each set of footprints, with line lengths proportionate to cleavage intensities. Not all data summarized in the schematics are from the portions of the gels shown. Ovals, inferred translational positions of nucleosomes; dashed rectangle, an MNase-resistant, DNase I-hypersensitive structure. HindIII designations pertain to the nuclear accessibility assay of Fig. 10. The HindIII site is at −274 in the WT allele and at −242 in the deletion allele.