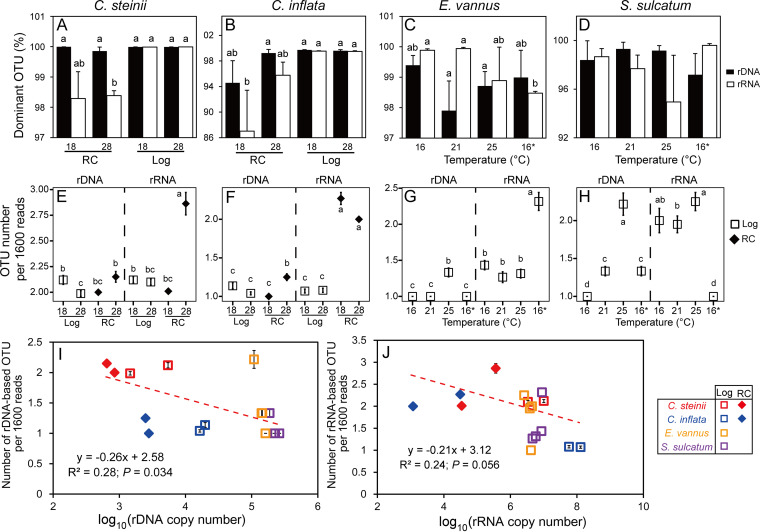
FIG 5.
Characterization of operational taxonomic units (OTUs) in rDNA and rRNA pools in single cells of four ciliate species. (A to H) Comparisons of the relative abundance of the most dominant OTU (A to D) and OTU numbers per 1,600 reads (E to H) between resting cysts (RC) and log-phase vegetative cells (Log) of Colpoda steinii and C. inflata at 18°C and 28°C and in a single log-phase cell of Euplotes vannus and Strombidium sulcatum at four temperatures. (I, J) Both rDNA (I) and rRNA-based OTU numbers (J) are negatively related to sequence number within a single cell. Two treatments not sharing common letters indicate significant differences (multiple pairwise comparisons, P < 0.05). Note that the OTUs are defined at a sequence similarity cutoff of 99% for C. steinii and C. inflata and at 100% for E. vannus and S. sulcatum based on the results of sequencing and analysis of individual clones with 18S rDNA or cDNA fragments.