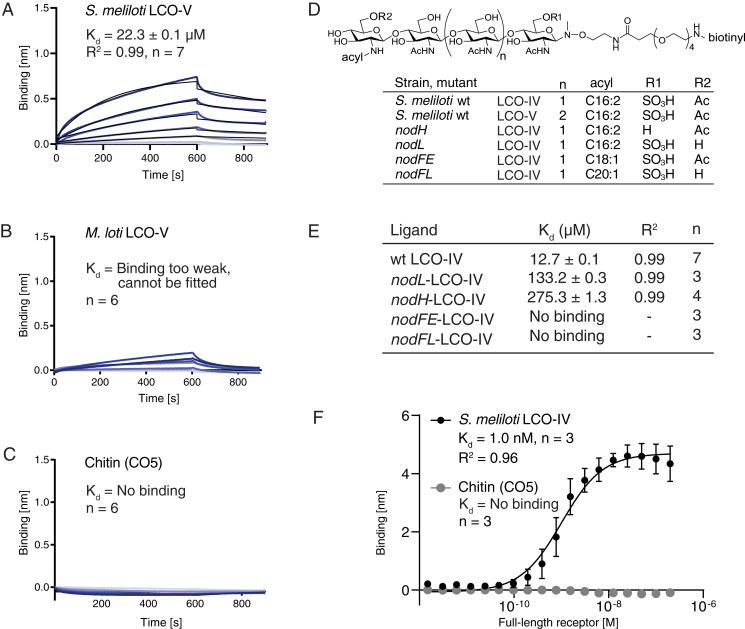
Fig. 2.
NFP has ligand specificity and directly monitors LCO decorations in BLI experiments. (A) NFP binding to S. meliloti LCO-V. (B) NFP binding to M. loti LCO-V is too weak and cannot be fitted. (C) NFP does not bind chitopentaose (CO5) in BLI experiments. A concentration range of analyte (100 to 1.56 µM) was used for each experiment. Experimental binding curves are represented in blue and fitting curves in black. The goodness of fit is described by the global fit R square of the mean value for each point. Numbers of replicates performed using independent protein preparations (n) are indicated. (D) Structure of biotinylated S. meliloti LCO-IV conjugate and overview of S. meliloti mutants associated with variations in LCO structure. S. meliloti LCO-IV has a tetrameric N-acetylglucosamine backbone, is O-sulfated on the reducing end, O-acetylated on the nonreducing terminal residue, and monoN-acylated by a hexadecadienoyl (C16:2) group. (E) BLI data showing NFP binding to S. meliloti LCO-IV variants. (F) Steady-state BLI data of full-length receptor binding at t = 595 s of association to immobilized LCO-IV and CO5, respectively. The binding follows a sigmoidal dose–response model with Kd = 1.0 ± 0.37 nM. Error bars indicate SD. A total of 16 twofold dilution series of analyte (200 to 0.0061 nM) were used for each experiment.