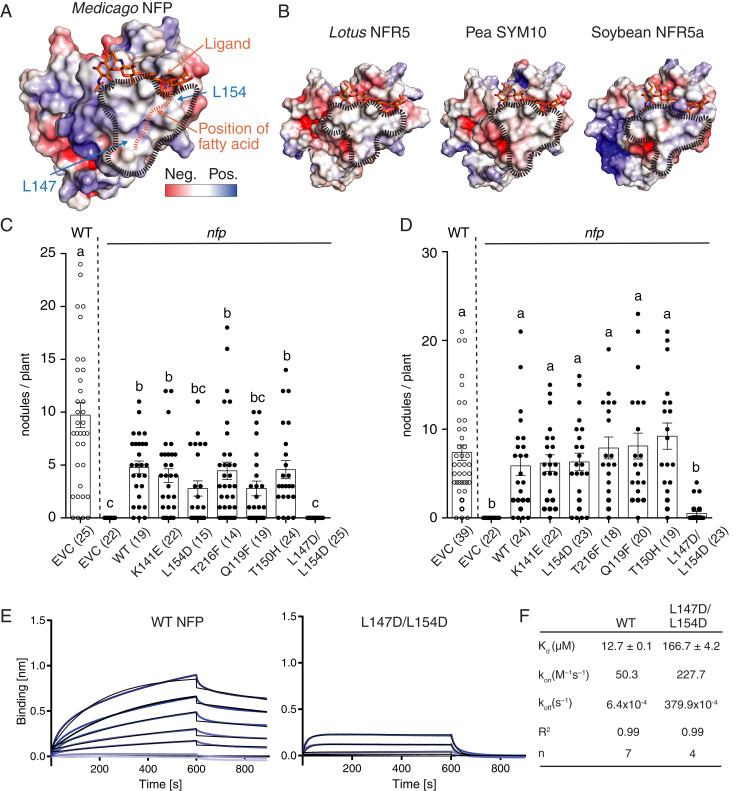
Fig. 3.
A hydrophobic patch in LysM2 is important for LCO binding and symbiotic signaling. (A) Molecular docking of a chitotetraose molecule (orange sticks) onto the structure of NFP. The surface of NFP is colored according to its electrostatic potential (±5 kT/e), and the hydrophobic patch is highlighted (black dashes). A possible position of the fatty acid chain on the hydrophobic patch is indicated (orange dashes). (B) Homology models of characterized LCO receptor ectodomains: L. japonicus NFR5, Pisum sativum (Pea) SYM10, and Glycine max (Soybean) NFR5α. All have a characteristic hydrophobic patch in LysM2. (C and D) Complementation analysis of NFP variants in an nfp Medicago background underlines that the hydrophobic patch is a prerequisite for functional symbiotic signaling. Columns represent mean nodule numbers after 49 d post infection (S. meliloti) (C) or 28 dpi (Sinorhizobium medicae) (D). Circles indicate individual counts. Empty circles: Medicago Jemalong wild-type background. Filled circles: nfp mutant background. EVC: empty vector control, WT: wild-type NFP. Error bars represent the SEM. Letters indicate statistical significance (ANOVA, Tukey, P < 0.05). Number of plants are indicated in parentheses. (E) BLI experiments of NFP WT and hydrophobic patch mutant (L147D/L154D) binding to S. meliloti LCO-IV. A concentration range of analyte (100 to 1.56 µM) was used for each experiment. Experimental binding curves are represented in blue and fitting curves in black. (F) Table summarizing the kinetic parameters for data in E. The goodness of fit is described by the global fit R square on the mean value of each point. Numbers of replicates performed using independent protein preparations (n) are indicated.