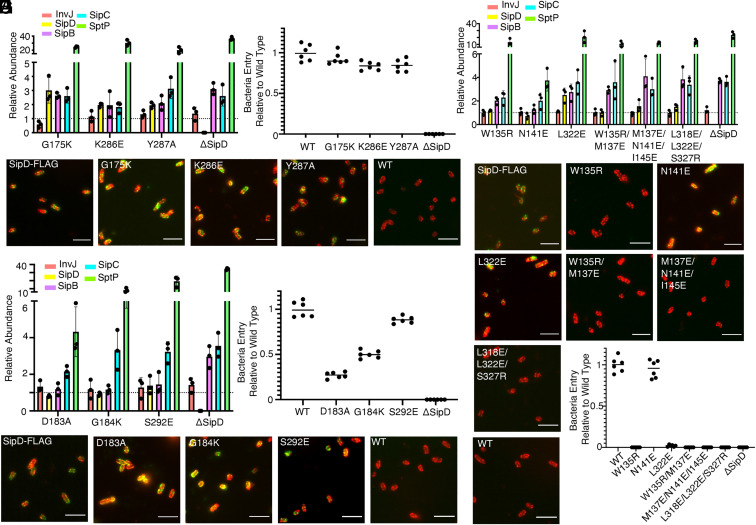
Fig. 6.
Structure and function analyses of the tip structure and its interface with the needle filament. (A–C) Functional analysis of S. Typhimurium strains expressing SipD variants with the indicated mutations in residues involved in the formation of the common SipD intersubunit interface. (A) Bar graph detailing the relative abundance of the indicated secreted substrates in culture supernatants standardized relative to the values of the WT strains (given a value of 1 and demarcated by a gray dashed line). All values represent the mean ± the SD of three independent experiments. The data were compiled from the data presented in Dataset S1. (B) Fluorescence microscopy images of intact, nonpermeabilized S. Typhimurium expressing FLAG-tagged wild-type SipD (SipD-FLAG), or the indicated mutant forms. Bacterial cells were probed with antibodies to the FLAG tag (green) and to lipopolysaccharide (red). The wild-type strain (with no tag) (WT) was included as a negative control. (Scale bars, 5 μm.) (C) Cultured epithelial cell invasion ability of S. Typhimurium strains expressing the indicated SipD mutants. Numbers represent the percentage of the inoculum that survived antibiotic treatment due to internalization and are the mean ± SD of six independent experiments normalized to wild type, which was set to 1. T3SS, type III secretion system; WT, wild type. (D) Secretion profile of different S. Typhimurium SipD1–SipD5 interface mutant strains shown by bar graph. The relative abundance of the secreted substrates has been standardized relative to the values of the WT strains, which were given a value of 1 and are demarcated by a gray dashed line. All values represent the mean ± the SD of three independent experiments. (D and E) Functional analysis of S. Typhimurium strains expressing SipD variants with the indicated mutations in residues involved in the formation of the SipD1–SipD5 interface. Secretion profiles (D), culture epithelial cell invasion (E), and fluorescence microscopy images (F) are shown. Experiments were conducted as indicated in A–C. (F–H) Functional analysis of S. Typhimurium strains expressing SipD variants with the indicated mutations in residues involved in the formation of the SipD–PrgI interface. Secretion profiles (G), fluorescence microscopy images (H), and culture epithelial cell invasion (I) data are shown. Experiments were conducted as indicated in A–C.