Figure 2.
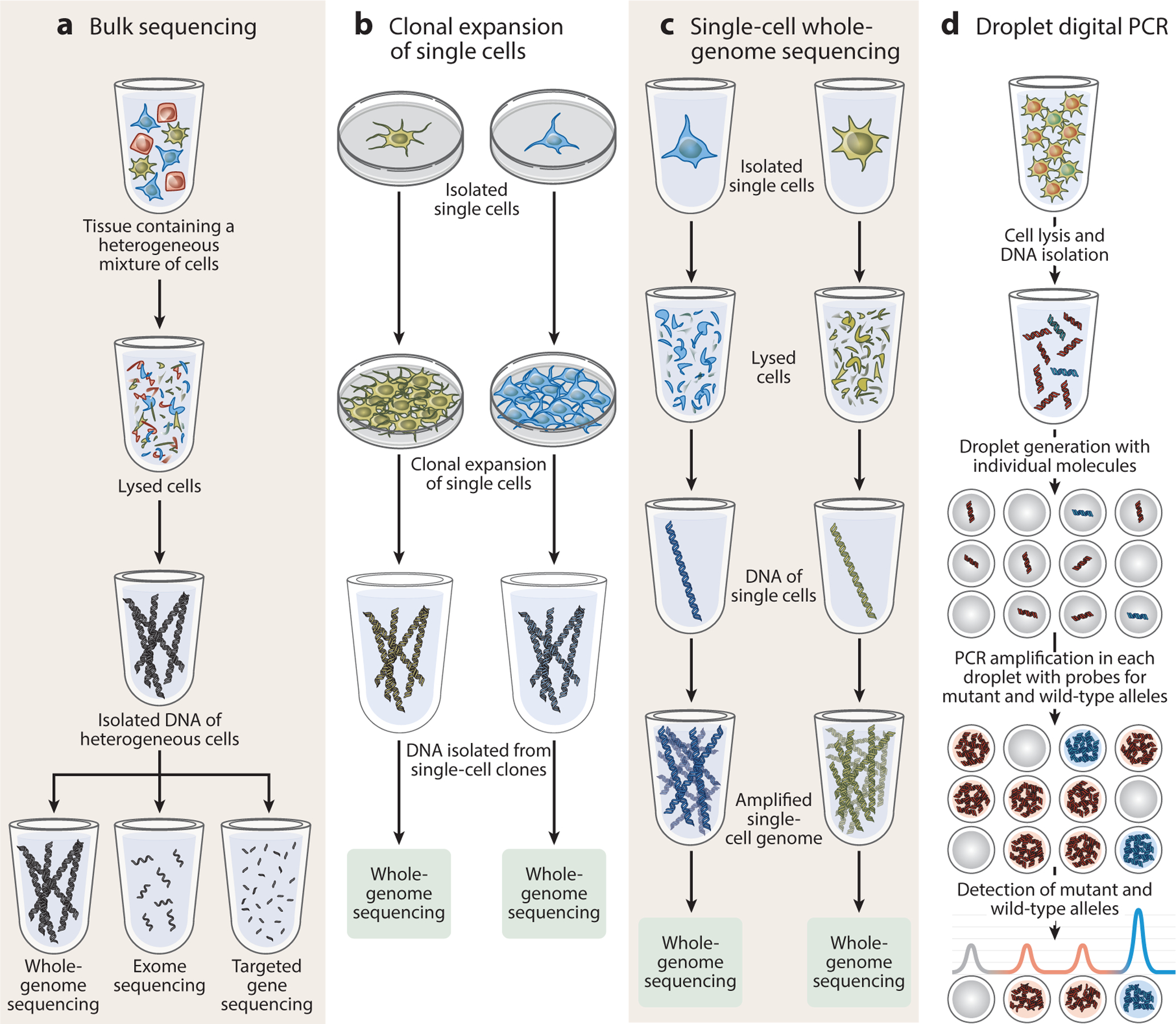
Examples of methods for detecting somatic mutations in bulk and single cells. The methods illustrated here focus on detecting somatic single-nucleotide variants, though some can also identify copy number variants and other classes of somatic mutation. (a) Bulk sequencing can be used to detect clonal somatic mutations, as they are present in multiple cells in a sample. DNA is isolated from a heterogeneous sample of cells, then used to prepare libraries for whole-genome sequencing or targeted sequencing methods, such as the exome or a specific set of genes. Bulk methods are used to detect variants arising during development, physiological proliferation, or neoplasia. Bulk sequencing is not well suited to detecting private mutations that are exclusive to individual cells, which generally require single-cell detection methods. However, bulk methods have higher throughput than current single-cell DNA sequencing methods, conveying information about more cells per experiment. (b) Clonal expansion of single cells in culture enables the biological amplification of the genome of a single initial cell, generating enough genome copies for detection by sequencing methods. Clonal expansion can be performed with natively proliferating cells or embryonic stem cells derived from nuclear transfer of any cell type. This method is susceptible to cell culture artifacts but minimizes in vitro genome amplification concerns. (c) Single-cell whole-genome sequencing facilitates the direct examination of the genome of a single target cell, typically using in vitro enzymatic genome amplification (e.g., by multiple displacement amplification). This technique can be used on any cell type (or even naked nuclei) that can be isolated using fluorescence-activated cell sorting or similar methods, and therefore can be applied to nondividing cells. (d) Variant-specific detection methods include droplet digital PCR. Bulk DNA is mixed with variant-specific primers and fluorescent probes, then fractionated into droplets. PCR amplification occurs on individual DNA fragments to produce a binary signal for each droplet and thus a quantitative measure of the variant allele fraction, and therefore this approach is often more sensitive than bulk sequencing methods. Figure created by Ken Probst of Xavier Studio, with input from the authors.
