Figure 8.
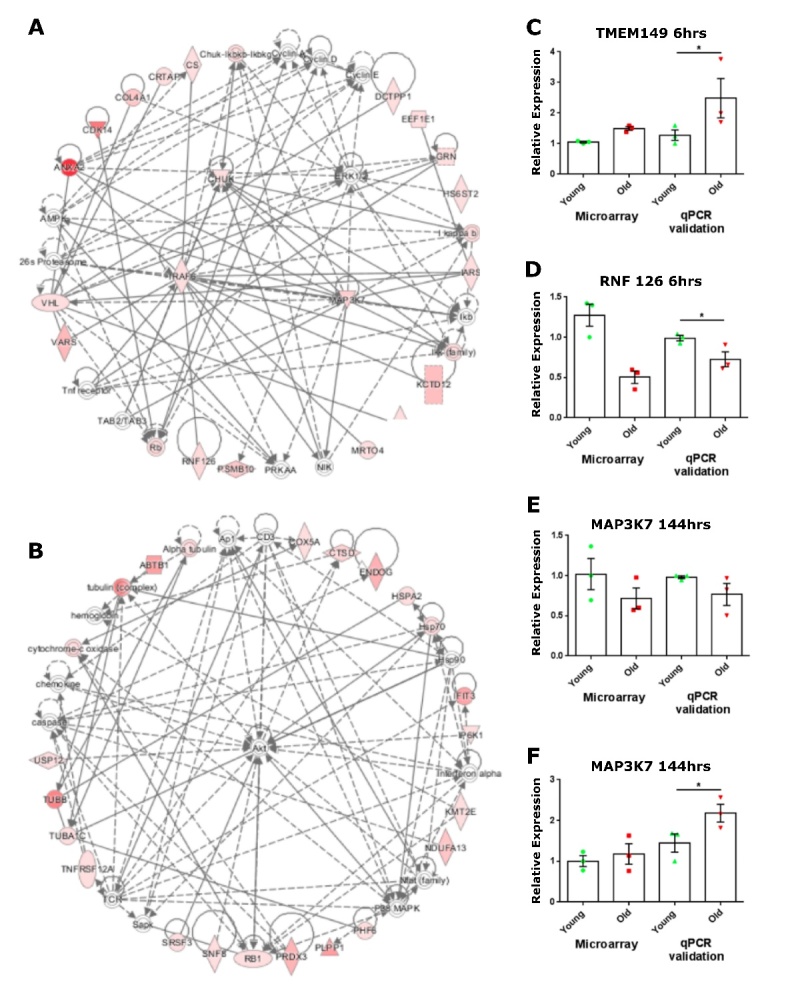
Validation of differentially expressed genes in response to serum from older adults. (A, B) Networks formed by differentially expressed genes in response to young and old serum. 111 differentially expressed genes during differentiation of hippocampal progenitors in the presence of young or old serum were analysed using Ingenuity Pathway Analysis (IPA) to assess network relationships between candidate genes, A) top network associated functions included “System Development and Function” and “Cell to Cell Signalling”, B) second highest network associated functions included “Cellular Development”, “Cellular Growth and Proliferation” and “Cancer”. (A, B) significance of genes reflected on continuous colour scale from red (most significant) to light pink (least significant). Inferred molecular interactions identified by IPA are shown in grey. (C-F) Validation by qPCR of differentially expressed microarray candidate genes in response to young or old serum. Relative expression of microarray candidates C) transmembrane 149 (TMEM149) D) ring finger protein 126 (RNF126) E) mitogen-activated protein kinase 7 (MAP3K7) F) endonuclease G (ENDOG) normalised to one young-serum induced readout, corresponding to 1 on the y axis for both the microarray and qPCR validation. Each green circle (young, n = 3) or red square (old, n = 3) represents expression values during differentiation of hippocampal progenitors in the presence of human serum at the stated time point. Microarray statistics not included. Unpaired student one-tailed t-test conducted on qPCR data, * P < 0.05, error bars = SEM.
