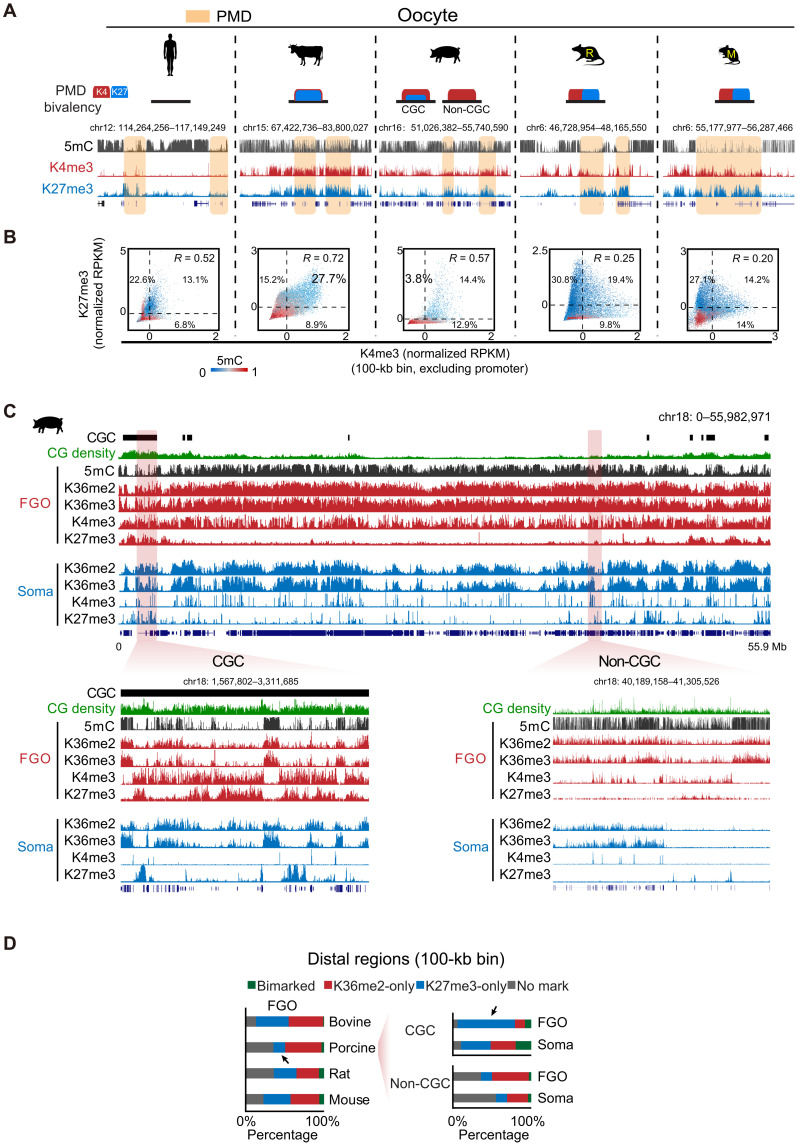
Fig. 5. Distribution of H3K4me3 and H3K27me3 and their relationship with H3K36me2/H3K36me3/DNA methylation in mammalian oocytes.
(A) The UCSC genome browser views showing the distribution of H3K4me3 and H3K27me3 and their relationship with DNA methylation in mammalian oocytes. The PMD regions are shaded. Schematic shows the states of bivalency (H3K4me3 and H3K27me3) in PMDs. (B) Scatterplots comparing distal H3K4me3 and H3K27me3 in each 100-kb bin (with Pearson correlation shown). DNA methylation levels in each bin are color-coded. The percentages of bins marked by both H3K4me3 and H3K27me3, H3K4me3-only, and H3K27me3-only are also shown in the plots. Note that bovine has high percentage of bivalent bins and porcine has very few H3K27me3-only bins. (C) The UCSC browser views showing H3K4me3, H3K27me3, and H3K36me2/3 in porcine oocytes. Histone marks in somatic CCs (soma) are shown as controls. Zoomed-in views of CGC and non-CGC are also shown. H3K27me3 is mainly enriched in CGCs. (D) Barcharts showing the percentages of the genome covered by H3K36me2 and H3K27me3 in the oocytes among different species (left). The percentages of genome covered by H3K36me2 and H3K27me3 in CGCs and non-CGCs are also shown for porcine FGOs (right). H3K27me3 shows limited overall coverage in the genome but shows high coverage in CGCs in porcine FGOs (arrows). The distribution in somatic CCs in porcine is calculated as controls.