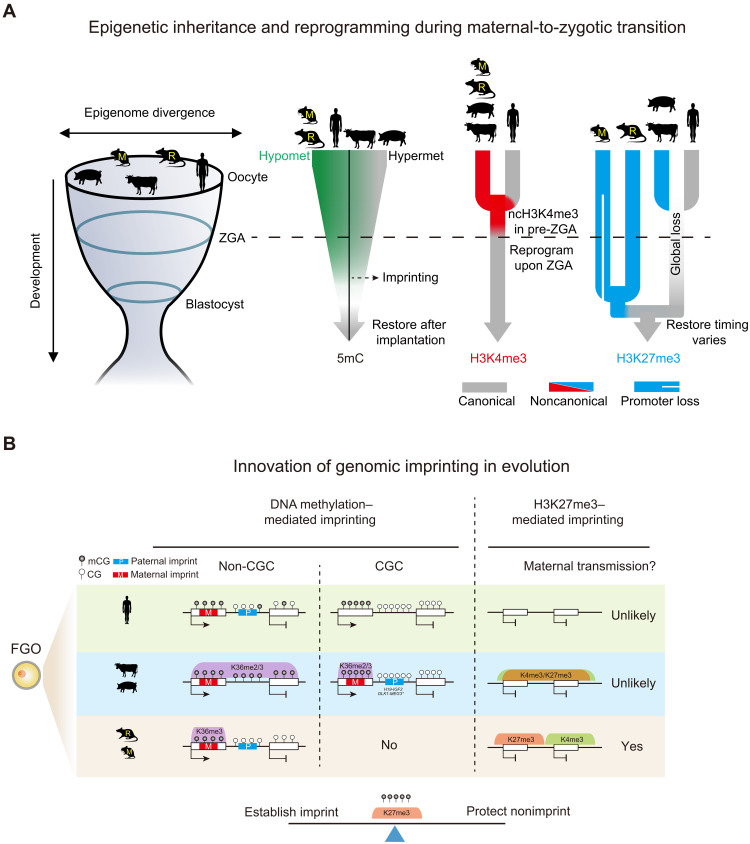
Fig. 8. Conservation and divergence of genomic imprinting and epigenetic reprogramming in mammalian early development.
(A) Schematic model showing the inheritance and reprogramming of DNA methylation, H3K4me3, and H3K27me3 during mammalian early development. Rodent oocytes show hypomethylated (Hypomet) DNA methylome while porcine oocytes show hypermethylated (Hypermet) DNA methylome. Human and bovine oocytes fall in between. After fertilization, DNA methylation is globally lost, except for genomic imprints, before being restored presumably after implantation. Oocyte ncH3K4me3 is conserved in nonhuman species. A “priming” state of H3K4me3 transiently appears in pre-ZGA human embryos (13). Both types of H3K4me3 are resolved to canonical patterns after ZGA. Broad ncH3K27me3 occurs in nonhuman oocytes. H3K27me3 is globally erased by the peri-ZGA stages in nonrodents. In rodents, promoter H3K27me3 is lost after fertilization in mouse but is partially retained in rat, while distal H3K27me3 is inherited until blastocysts. Embryo H3K27me3 is restored at variable stages (morula for human, blastocyst for bovine, and beyond blastocyst for porcine and rodents). DNA methylation, H3K4me3, and H3K27me3 show more similar patterns around implantation among species, supporting the “hourglass model” (left) (33). (B) Schematic model showing distinct mechanisms of genomic imprinting. DNA methylation–mediated imprinting is largely conserved among species. H3K36me2/3 correlated DNA methylation likely leads to the establishment of maternal imprints in the oocytes. Paternal imprints reside in nontranscribed regions. Some reside in CGCs (H19-IGF2 ICR for bovine, and H19-IGF2 and DLK1-MEG3 ICRs for porcine). Asterisk, DLK1-MEG3 ICR is not in CGC in bovine but is surrounded by CGCs. Oocyte-derived ncH3K27me3 can persist beyond ZGA and likely mediate noncanonical imprinting in rodents but unlikely in other species. Coincidently, ncH3K4me3 and ncH3K27me3 domains are segregated only in rodents. Oocyte finely balances the establishment of genomic imprints and the protection of nonimprints.