Abstract
In modern scientific research, data are often collected from multiple modalities. Since different modalities could provide complementary information, statistical prediction methods using multi-modality data could deliver better prediction performance than using single modality data. However, one special challenge for using multi-modality data is related to block-missing data. In practice, due to dropouts or the high cost of measures, the observations of a certain modality can be missing completely for some subjects. In this paper, we propose a new DIrect Sparse regression procedure using COvariance from Multi-modality data (DISCOM). Our proposed DISCOM method includes two steps to find the optimal linear prediction of a continuous response variable using block-missing multi-modality predictors. In the first step, rather than deleting or imputing missing data, we make use of all available information to estimate the covariance matrix of the predictors and the cross-covariance vector between the predictors and the response variable. The proposed new estimate of the covariance matrix is a linear combination of the identity matrix, the estimates of the intra-modality covariance matrix and the cross-modality covariance matrix. Flexible estimates for both the sub-Gaussian and heavy-tailed cases are considered. In the second step, based on the estimated covariance matrix and the estimated cross-covariance vector, an extended Lasso-type estimator is used to deliver a sparse estimate of the coefficients in the optimal linear prediction. The number of samples that are effectively used by DISCOM is the minimum number of samples with available observations from two modalities, which can be much larger than the number of samples with complete observations from all modalities. The effectiveness of the proposed method is demonstrated by theoretical studies, simulated examples, and a real application from the Alzheimer’s Disease Neuroimaging Initiative. The comparison between DISCOM and some existing methods also indicates the advantages of our proposed method.
Keywords: Block-missing, Huber’s M-estimate, Lasso, Multi-modality, Prediction, Sparse regression
1. Introduction
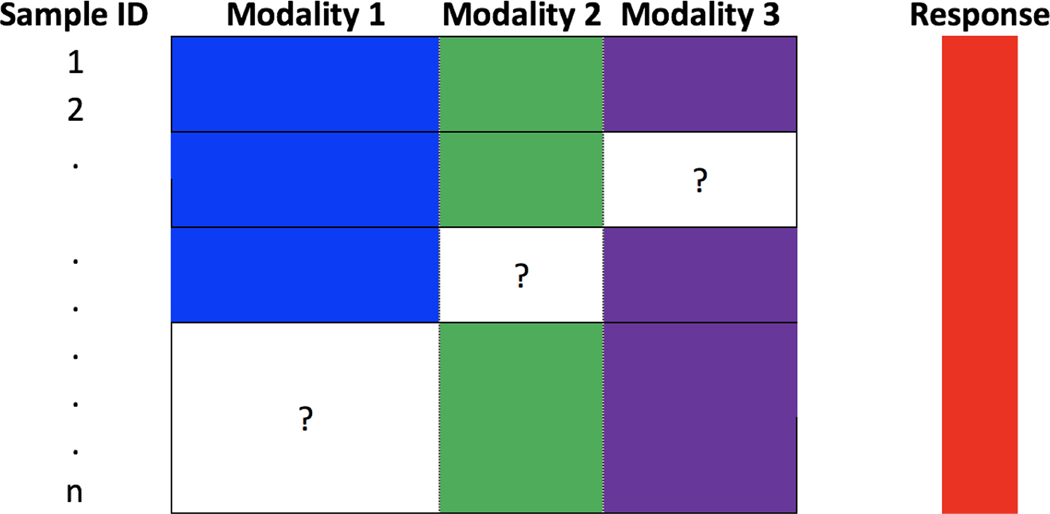
With the advance of modern scientific research, complex data are often collected from multiple modalities (sources or types). In neuroscience, different brain images such as magnetic resonance imaging (MRI) and positron emission tomography (PET) are used to study the brain structure and function. In biology, data from different modalities such as gene expressions and copy numbers are collected to understand the complex mechanism of cancers. Since different modalities could provide complementary information, statistical prediction methods using multi-modality data could deliver better prediction performance than using single modality data. However, one special challenge for using multi-modality data is related to missing data, which is unavoidable due to some reasons such as the high cost of measures or the patients’ dropout. Generally, the observations of a certain modality can be missing completely, i.e., a complete block of the data is missing. One example of block-missing multi-modality data is shown in Figure 1. In this example, there are n samples (each row represents one sample), three modalities and one response variable. The blank regions with question mark indicate missing data. As shown in Figure 1, for many samples, the observations from some modality are missing completely. The number of samples with complete observations is much smaller than the sample size n.
Figure 1:
An illustration of block-missing multi-modality data with three modalities.
To predict the response variable using the high dimensional block-missing multi-modality data, a common strategy is to use the Lasso (Tibshirani (1996)) or some other penalized regression methods (e.g., Fan and Li (2001); Zou and Hastie (2005); Zhang (2010)) only for the data with complete observations. However, this strategy can greatly reduce the sample size and waste a lot of useful information in the samples with missing data. Another strategy is to impute the missing data first by some existing imputation methods (Hastie et al. (1999); Cai et al. (2010)). These methods can be effective when the positions of the missing data are random, but they can be unstable when a complete block of the data is missing. Recently, motivated by applications in genomic data integration, Cai et al. (2016) proposed a new framework of structured matrix completion to impute block-missing data. However, they only consider the case when the data are collected from two modalities. In the literature, rather than deleting or imputing missing data, some studies focus on using all available information. For example, Yuan et al. (2012) proposed the Incomplete Multi-Source Feature learning (IMSF) method. The IMSF method performs regression on block-missing multi-modality data without imputing missing data. It formulates the prediction problem as a multi-task learning problem by first decomposing the prediction problem into a set of regression tasks, one for each combination of available modalities (e.g., modalities 1, 2 and 3; modalities 1 and 2; modalities 1 and 3; modalities 2 and 3 for the example shown in Figure 1), and then building regression models for all tasks simultaneously. The important assumption in the IMSF method is that all models involving a specific modality share the common set of predictors for that particular modality. However, when different modalities are highly correlated, this assumption could be too strong. In that case, for some modalities, it can be more reasonable to choose different predictor subsets for different involved tasks. Therefore, it is desirable to develop flexible and efficient prediction methods applicable to block-missing multi-modality data.
In this paper, we propose a new DIrect Sparse regression procedure using COvariance from Multi-modality data (DISCOM). For each sample, if some modality has missing entries, all the observations from that modality are missing simultaneously. Regardless of the underlying true model, we aim to find the optimal linear prediction for the response variable using the block-missing multi-modality data without imputing the missing data. Our method includes two steps. In the first step, we use all available information to estimate the covariance matrix of the predictors and the cross-covariance vector between the predictors and the response variable. The proposed new estimate of the covariance matrix is a linear combination of the identity matrix, the estimates of the intra-modality covariance matrix and the cross-modality covariance matrix. Flexible estimates for both the sub-Gaussian and heavy-tailed cases are considered. Many existing high dimensional covariance estimation methods such as Bickel and Levina (2008); Cai and Liu (2011); Rothman (2012); Lounici et al. (2014); Cai and Zhang (2016) can be used in this step. In the second step, based on the estimated covariance matrix and the estimated cross-covariance vector, we use an extended Lasso-type estimator to estimate the coefficients in the optimal linear prediction.
Note that there are some existing sparse regression methods in the literature using the estimation of the covariance matrix. For example, Jeng and Daye (2011) proposed the covariance-thresholded Lasso for complete data to improve variable selection by utilizing the sparsity of the covariance matrix. Loh and Wainwright (2012) and Datta et al. (2017) proposed new estimators for the high dimensional regression with corrupted predictors, where all entries of the design matrix are assumed to be noisy or missing randomly and independently. The missing data problem they considered can be viewed as a special case of the block-missing multi-modality data where each modality has only one predictor. To the best of our knowledge, there are no existing methods using a similar idea to DISCOM tailored for high-dimensional block-missing multi-modality data. To investigate DISCOM, we have carefully studied its theoretical and numerical performance. For both the sub-Gaussian and heavy-tailed cases, we establish the consistency of estimation and model selection for the optimal linear predictor regardless of the underlying true model. Our theoretical studies indicate that DISCOM could make use of all available information of the block-missing multi-modality data effectively. The number of samples that are effectively used by DISCOM is the minimum number of samples with available observations from two modalities, which can be much larger than the number of samples with complete observations from all modalities. The comparison between DISCOM and some existing methods using simulated data and the Alzheimer’s Disease Neuroimaging Initiative (ADNI) data (www.loni.ucla.edu/ADNI) further demonstrate the effectiveness of our proposed method.
The rest of this paper is organized as follows. In Section 2, we motivate and introduce our method. In Section 3, we show some theoretical results about the estimates of the covariance matrix, the cross-covariance vector and the coefficients in the optimal linear prediction for both the sub-Gaussian and heavy-tailed cases. The results about the model selection consistency are also provided. In Sections 4 and 5, we demonstrate the performance of our method on the simulated data and the ADNI dataset. We conclude this paper in Section 6 and provide all technical proofs in the Appendix.
2. Motivation and Methodology
We first show the motivation and the outline of our proposed method in Section 2.1. In Section 2.2, we introduce the proposed estimate of the covariance matrix of the predictors, and the estimate of the cross-covariance vector between the predictors and the response variable using the block-missing multi-modality data. In Section 2.3, we introduce the Huber’s M-estimate for the heavy-tailed case. In Section 2.4, we provide the estimation procedure for the coefficients in the optimal linear prediction.
The following notation will be used in this paper. For a matrix A ∈ Rm×n, we use , , and , to denote the Frobenius norm , the max norm maxij |aij|, and the infinity norm , respectively. For a vector b ∈ Rm×1, we use ∥b∥2, ∥b∥max, and ∥b∥1 to denote the norm , the max norm maxi |bi|, and the norm , respectively. In addition, we use sign(·) to denote the function that maps a positive entry to 1, a negative entry to −1, and 0 to 0.
2.1. Motivation
Suppose the predictors are collected from K modalities. For , there are pk predictors from the k-th modality. Let n denote the sample size, denote the n × 1 response vector centered to have mean 0, and denote the design matrix of the pk predictors from the k-th modality. In addition, let denote the n × p design matrix, where . We assume that xi’s are i.i.d. generated from some multivariate distribution with mean 0p×1 and covariance matrix Σ. We use to denote the cross-covariance vector between xi and yi.
To predict the response variable y using all predictors X1, X2, . . ., Xp, we consider the optimal linear predictor , where the coefficient vector
| (1) |
The above coefficient vector β0 can be viewed as the solution to the following optimization problem
If we know the true covariance matrix Σ and the true cross-covariance vector C, and assume that β0 is sparse, we can estimate β0 by solving the following optimization problem
| (2) |
where λ is a nonnegative tuning parameter.
Motivated by (2), for the high dimensional block-missing multi-modality data, we propose a new method with two steps. In the first step, we use all available observations to estimate the covariance matrix Σ and the cross-covariance vector C. The estimates of Σ and C are denoted as and , respectively. This step is very important to make full use of the block-missing multi-modality data. In the second step, we estimate β0 by solving the following optimization problem:
| (3) |
2.2. Standard estimates of Σ and C
Considering block-missing multi-modality data, for each sample, if a certain modality has missing entries, all the observations from that modality are missing. For each predictor j, define Sj = {i : xij is not missing}. For each pair of predictors j and t, define Sjt = {i : xij and xit are not missing}. The number of elements in Sj and Sjt are denoted as nj and njt, respectively.
For the missing data mechanism, we only need to assume that for each predictor, the first sample moment and the second sample moment using all available observations are unbiased estimators of the first theoretical moment and the second theoretical moment of the distribution, respectively. This assumption is satisfied if we assume that each modality is missing completely at random. However, different predictors in the same modality are missing simultaneously. Under this assumption, for each j ∈ {1, 2, . . ., p}, the available observations of the j-th predictor are centered to have mean 0. A natural initial unbiased estimate of Σ using all available data is the sample covariance matrix
For the block-missing multi-modality data, the above initial estimate may have negative eigenvalues due to the unequal sample sizes njt’s. Therefore, it is not a good estimate of the covariance matrix Σ and not suitable to be used in (3) directly. It is important to find an estimator that is both positive semi-definite and more accurate than the initial estimate .
According to the partition of the predictors into K modalities, the initial estimate of the covariance matrix can be partitioned into K2 blocks, denoted by ’s, where j, t ∈ {1, 2, . . ., p} and is a pj × pt matrix. We donte
where is called the intra-modality sample covariance matrix which is a p × p block-diagonal matrix containing K main diagonal blocks of , and is called the cross-modality sample covariance matrix containing all the off-diagonal blocks of . We also let ΣI and ΣC denote the true intra-modality covariance matrix and cross-modality covariance matrix, respectively. As shown in Figure 1, since the observations of some modalities are missing completely for many samples, there are more available samples to estimate the intra-modality covariance matrix ΣI than the cross-modality covariance matrix ΣC. Intuitively, it is relatively easier to estimate ΣI than ΣC. In view of this characteristic of the block-missing multi-modality data and the possible negative eigenvalues of , we propose to use the following estimator
where α1, α2 and α3 are three nonrandom weights, and Ip is a p × p identity matrix. Considering all possible linear combinations, we can find the optimal linear combination whose expected quadratic loss is the minimum. The optimal weights , and are shown in the following Proposition 1. As a remark, Proposition 1 and all the theoretical analysis in Section 3 are conditional on the given missing pattern of different modalities.
Proposition 1.
Consider the following optimization problem:
where the weights α1, α2 and α3 are nonrandom. Denote , , , and . The optimal weights are
In addition, we have
Proposition 1 shows that is more accurate than . The relative improvement in the expected quadratic loss over the sample covariance matrix is equal to
Therefore, if is relatively accurate ( is small), the optimal weight should be large and the percentage of the relative improvement tends to be small. We can also make the same conclusions about . For the block-missing multi-modality data, due to the unequal sample sizes, the initial estimate can be relatively accurate while the estimate is relatively inaccurate. It’s reasonable to use different weights for and . As a remark, Proposition 1 can be viewed as a generalization of Theorem 2.1 shown in Ledoit and Wolf (2004), where they studied the optimal linear combination of the sample covariance matrix and the identity matrix to estimate the covariance matrix for the complete data.
Regarding the cross-covariance vector C, we can use the following estimate
Note that we use all available information to estimate Σ and C. The theoretical properties of and will be discussed in Section 3.
2.3. Robust estimates of Σ and C
When the predictors and the response variable follow a sub-Gaussian distribution with an exponential tail, and introduced in Section 2.2 generally perform well. However, when the distributions of the predictors and the response variable are heavy-tailed, and may have poor performance, and therefore some robust estimates of Σ and C are required.
In this section, we introduce robust estimates of Σ and C based on the Huber’s M-estimator (Huber et al. (1964)). In general, suppose Z1, Z2, . . ., Zn are i.i.d. copies of a random variable Z with mean μ. The Huber’s M-estimator of μ is defined as the solution to the following equation
where is the Huber function which is given by
Using the Huber’s M-estimator, for the block-missing multi-modality data, we can construct a robust initial estimate of Σ denoted by
In general, the parameters Hjt used in the Huber function can be chosen to be 1.345 in order to guarantee 95% efficiency relative to the sample mean if the data generating distribution is Gaussian (Huber et al. (1964)). However, for the block-missing multi-modality data, considering different numbers of samples available to estimate different entries of Σ, we propose to use different values of H flexibly. The choice of Hjt will be discussed in Section 3. Based on the robust initial estimate , we can use a similar idea introduced in Section 2.2 to find the optimal linear combination whose expected quadratic loss is the minimum. Similarly, we can use the Huber’s M-estimator to deliver a robust estimate of C which is defined as
Here we also propose to use different values of H when estimating different cj’s. The choice of Hj will be discussed in Section 3. The theoretical properties of and will be also shown in that section.
2.4. Estimate of β0 in the optimal linear prediction
After getting an initial estimate of Σ and C, e.g., and (or and ), our proposed DISCOM method estimates β0 by solving the following optimization problem:
| (4) |
where , are two weights and tr()/p is used to estimate γ*. In practice, both , , and λ can be chosen by cross validation or an additional tuning dataset. To guarantee that the estimated covariance matrix is positive semi-definite, we need to choose reasonable α1 and α2 from the set , where is the smallest eigenvalue of .
Besides the above tuning parameter selection method that searches for the best values of three parameters, we can use an efficient tuning method incorporating our theoretical results in Section 3. Our theoretical studies show that the tuning parameters α1 and α2 should satisfy the conditions and , respectively. Denote and . We can choose α1 = 1 − k0m1 and α2 = 1 − k0m2, where k0 ∈ [kmin, kmax] is a tuning parameter. To guarantee that both α1 and α2 are nonnegative, we set kmax = min{1/m1, 1/m2}. In addition, a reasonable value of k0 should satisfy the following two conditions: (1) α1 = 1 − k0m1 ≤ 1 and α2 = 1 − k0m2 ≤ 1; (2) the estimate of the covariance matrix is positive semi-definite. The first condition requires that k0 ≥ 0. If the smallest eigenvalue of the initial estimate , denoted by λmin(), is nonnegative, we can show that is positive semi-definite for any nonnegative k0. If λmin() < 0, since the smallest eigenvalue of satisfies
to guarantee that is positive semi-definite, we only need to require that
Therefore, if , we choose kmin = 0. Otherwise, we choose
For the block-missing multi-modality data, since m2 ≥ m1 > 0, we know that the matrix is positive definite and therefore kmin is always less than .
By choosing α1 = 1 − k0m1 and α2 = 1 − k0m2, our proposed fast tuning parameter selection method searches the best value of k0 ∈ [kmin, kmax] and the parameter λ rather than searching three parameters α1, α2 and λ. In addition, instead of using the eigendecomposition for each parameter combination to check whether is positive semi-definite, this method only requires two eigendecompositions of the matrices and before the tuning parameter selection process. For each k0 ∈ [kmin, kmax], we can incorporate the coordinate descent algorithm (Friedman et al. (2010)) on a grid of λ values, from the largest one down to the smallest one, using warm starts. Alternatively, since is positive semi-definite, we can use the LARS algorithm shown in Jeng and Daye (2011) to compute the solution path.
As many existing high dimensional linear regression studies for the random design, we use the assumption E(X) = 0 to make our presentation more convenient. Our proposed DISCOM method can be used for the general case where E(X) ≠ 0. In that case, we first center the available observations of each predictor and use to denote the sample means of those p predictors. We also center the observed responses and use Ȳ to denote the sample mean of the response variable. Let denote the estimated regression coefficient vector calculated from the centered data. Our final predictive model is , where is a test data point. In practice, if our data are collected at various time points by different laboratories using multiple platforms, the i.i.d. assumption may be violated due to batch-effects. In that case, we suggest to use some existing statistical methods (e.g., the exploBATCH R package) to diagnose, quantify and correct batch effects before using our proposed DISCOM method.
3. Theoretical Study
Without loss of generalization, we assume that the true variances of all predictors, σ11, σ22, . . ., σpp, are equal to 1 in our theoretical studies. For each j ∈ {1, 2, . . ., p}, we assume that the observations of the predictor j are scaled such that . In that case, we have . For the Huber’s M-estimator , we redefine to be 1 for each j. Let and denote the solutions to (4) using the sample covariance and the Huber’s M-estimator, respectively. We assume that β0 is sparse and denote as the index set of the important predictors. Denote as the number of important predictors. Let and . In Sections 3.1 and 3.2, we will discuss the theoretical properties in the sub-Gaussian case and the heavy-tailed case, respectively. The model selection consistency of our proposed method will be shown in Section 3.3.
3.1. Sub-Gaussian case
The following conditions are considered in this section:
(A1) Suppose that there exists a constant L > 0 such that
(A2) Suppose that the true covariance matrix Σ satisfies the following restricted eigenvalue (RE) condition
Under condition (A1), the predictors and the response variable follow sub-Gaussian distributions with exponentially bounded tails. In this case, we propose to use and shown in Section 2.2 as the initial estimate of the covariance matrix Σ and the cross-covariance vector C, respectively. The RE condition (A2) is often used to obtain bounds of statistical error of the Lasso estimate (Datta et al. (2017)). The following Theorem 1 shows the large deviation bounds of and .
Theorem 1.
Under condition (A1), if minj,t njt ≥ 6 log p, there exists two positive constants ν1 = 8(1 + 4L2) and ν2 = 4 such that
There exists another two positive constants and ν4 = 4 such that
Remark 1.
In our theoretical studies, we assume that the dimension p goes to infinity as the sample size minj,t njt increases. If we further assume that (log p)/minj,t njt = o(1), the condition minj,t njt > 6 log p is satisfied if the sample size minj,t njt is sufficiently large. Then, Theorem 1 shows that . The performance of depends on the worst case when there are only minj,t njt samples to estimate some entries in Σ. In addition, the convergence rate of is . The performance of also depends on the worst case when there are only minj nj samples to estimate the covariance between some predictor and the response variable. Furthermore, if we only use samples with complete observations, using a similar proof, we can show that and , where ncomplete is the number of samples with complete observations. For the block-missing multi-modality data, since ncomplete can be much smaller than minj,t njt and minj nj, Theorem 1 indicates that the first step of our proposed DISCOM method can make full use of all available information. Based on the results shown in Theorem 1, we will show the convergence rate of .
Theorem 2.
Under conditions (A1) and (A2), let and . If and we choose , then we have .
Remark 2.
As shown in the above Theorem 2, we have . If we assume that (a) there is no missing data, (b) the predictors are generated from a multivariate Gaussian distribution, and (c) the true model is Y = Xβ0 + ϵ, where ϵ ∼ N(0, σ2In). Then we will use and to estimate Σ and C, respectively. Therefore, we have , and , which is the minimax ℓ2-norm rate as shown in Raskutti et al. (2011). Since the complete data generated from the Gaussian random design can be viewed as a special type of block-missing multi-modality data, the error bound in Theorem 2 is sharp.
On the other hand, if the true relationship between the conditional expectation and the predictors is non-linear, we have and as shown in the proof. In this case, if we still use the Lasso method to estimate the regression coefficients β0 in the optimal linear predictor, we have . For the blocking missing multi-modality data, since the Lasso method can only use the data with complete observations, we have . However, as shown in Theorem 2, for our proposed DISCOM estimate , we have . In practice, the minimum number of samples with available observations from two modalities (minj,t njt) can be much larger than the number of samples with complete observations from all modalities (ncomplete). Theorem 2 indicates that DISCOM could make use of the block-missing multi-modality data more effectively than the Lasso method using only the complete data.
In Theorem 2, the assumption is used to guarantee that satisfies the RE condition with a high probability if the true covariance matrix Σ satisfies the RE condition (A2). Note that many existing sparse linear regression studies focus on the fixed design where the design matrix X is considered to be fixed and complete. In that case, is assumed to satisfy the RE condition directly. For the general random design, Van De Geer et al. (2009) showed that satisfies the RE condition as long as the true covariance matrix satisfies the RE condition and s2 logp/n = o(1). For the special Gaussian random design, by a global analysis of the full random matrix rather than a local analysis looking at individual entries of , Raskutti et al. (2010) shows that the matrix satisfies the RE condition with a high probability if the true covariance matrix of the multivariate Gaussian distribution satisfies the RE condition and n > Constant·s log p. In our paper, since we consider the general random design including both sub-Gaussian distributions and heavy-tailed distributions, and study the proposed estimated covariance matrix where in most cases, we use the condition to guarantee that the RE condition is satisfied with a high probability. This condition is very similar to the condition used in Van De Geer et al. (2009) for the complete data.
For the general random design and the block-missing multi-modality data, it is difficult to develop a weak condition (e.g., s log p/minj,t njt = o(1)) using a similar global analysis of the full random matrix as shown in Raskutti et al. (2010). Instead of using the condition , we can use the following weak condition
where is a positive constant. This condition is also used in some existing studies about random designs (Bühlmann and Van De Geer (2011); Zhou et al. (2009)).
3.2. Heavy-tailed case
In this section, we consider the heavy-tailed case. Instead of assuming that the distributions of the predictors and the response variable have exponential tails, we consider the following moment condition.
(A3) Suppose that max and , where Q1 and Q2 are two positive constants.
Condition (A3) assumes that the fourth moments of all predictors Xj’s and the response variable y are bounded. Under condition (A3), the tails of the distributions of Xj’s and y may not be exponentially bounded. In the literature on Lasso, most studies consider the fixed design (Zhao and Yu (2006); Zou (2006); Meinshausen and Bühlmann (2006)) and the noise is usually assumed to be Gaussian (Meinshausen and Bühlmann (2006); Zhang et al. (2008)), or admits exponentially bounded tail (Bunea et al. (2008); Meinshausen and Yu (2009)). In this study, we consider a random design case and relax the distribution of Xj’s and y to have finite fourth moments.
Next, we discuss the theoretical properties of the Huber’s M-estimators and . Based on the convergence rates of and , we will show the convergence rate of .
Theorem 3.
Under condition (A3), let for each j, t ∈ {1, 2, . . ., p}, if minj,t njt ≥ 24 log p, we have
In addition, let for each , we have
Remark 3.
If we assume that (log p)/minj,t njt = o(1), the condition minj,t njt > 24 log p is satisfied if the sample size minj,t njt is sufficiently large. Therefore, we have and . This indicates that the Huber’s M-estimators for the heavy-tailed case acquire the same convergence rate as the sample covariance estimates for the sub-Gaussian case. However, as shown in the next theorem, if the distributions of the predictors Xj’s and the response variable y are not assumed to have exponentially bounded tails, the large deviation bounds of and can be wider than the bounds of the Huber’s M-estimators and , respectively.
Theorem 4.
Suppose and , where T > 0, ℓ > 1 are two constants. Then we have
where d1 > 0, d2 > 0, h ∈ (1, ℓ) are some constants. Furthermore,
where d3 > 0 and d4 > 0 are two constants.
Remark 4.
Under the moment condition, Theorem 4 shows that and . According to the Proposition 6.2 in Catoni (2012), the bounds shown in Theorem 4 are actually tight. If the dimension p is very large, the large deviation bounds of and can be much larger than the bounds of and , respectively. This necessitates the usage of a robust estimator.
In the next theorem, based on the large deviation bounds of and , we show the convergence rate of .
Theorem 5.
Under conditions (A2) and (A3), let , , and . If and let , then we have .
Remark 5.
Instead of using the condition = o(1), we can assume that
where is a positive constant. Theorem 5 indicates that for the heavy-tailed case, under (A3), the convergence rate of is also , which is the same as the rate shown in Theorem 2 under the sub-Gaussian assumption. However, as shown in our simulation study, if the response variable and the predictors follow sub-Gaussian distributions, DISCOM using standard estimates and generally has better finite sample performance than the method using robust estimates and .
Remark 6.
If we assume that p is fixed, for the sub-Gaussian case considered in Section 3.1, we can show that and according to Lemma 1 in Ravikumar et al. (2011) and a very similar proof of Theorem 1. For the heavy-tailed case considered in Section 3.2, if we assume that p is fixed, we can also show that and according to Theorem 5 in Fan et al. (2016) and a very similar proof of Theorem 3. Then, using the same proof of Theorem 2, we can also show that . Since , , and p is fixed, we can further show that . Similarly, for the heavy-tailed case, we can also show that . Therefore, the convergence rate of the estimation error in the classical fixed p setting is faster than the rate in the high dimensional setting where p grows to infinity.
3.3. Model selection consistency
In this section, we show that our proposed DISCOM method is model selection consistent. The following condition is considered.
(A4) , where η ∈ (0, 1) is a constant, is the sub-matrix of Σ with row indices in the set Jc and column indices in the set J, and ΣJJ is the sub-matrix of Σ with both row and column indices in the set J.
Condition (A4) can be viewed as a population version of the strong irrepresentable condition proposed in Zhao and Yu (2006). In the following Theorem 6 and Theorem 7, we will show that our proposed DISCOM method is model selection consistent for the sub-Gaussian case and the heavy-tailed case, respectively.
Theorem 6.
Under conditions (A1) and (A4), let and . If , and
then there exists a solution to (4) such that , as minjt njt → ∞ and p → ∞.
Remark 7.
Note that the condition is used to guarantee that (a) and (b) if for the general random design with a high probability, where and η ∈ (0, 1) are two constants. For the fixed design, we do not need this condition. For the special Gaussian random design, as shown in Wainwright (2009), using some concentration inequalities about the normal distribution and the fact that for the complete data, we can obtain model selection consistency with n > Constant · s log(p − s). In our theoretical studies, since we consider the general random design including both sub-Gaussian distributions and heavy-tailed distributions, and for the block-missing multi-modality data, we use the condition to guarantee that (a) and (b) are satisified. Note that this condition was also used in some existing model selection consistency studies for random designs (Jeng and Daye (2011); Datta et al. (2017)).
As shown in the proof of Theorem 6, to guarantee that (a) and (b) are satisfied, instead of requiring , we can use the following weak condition
where is a positive constant.
Theorem 7.
Under conditions (A3) and (A4), let , , , . If , and
then there exists a solution to (4) such that , as and .
Remark 8.
Instead of requiring , we can use the following weak condition
where is a positive constant. The proof of Theorem 7 is very similar to the proof of Theorem 6. We only show the proof of Theorem 7 briefly in the Appendix.
4. Simulation Study
In this section, we perform numerical studies using simulated examples. We use DISCOM and DISCOM-Huber to denote our proposed methods using sample covariance estimates and Huber’s M-estimates, respectively. The proposed methods using the fast tuning parameter selection method shown in Section 2.4 are called Fast-DISCOM and Fast-DISCOM-Huber, respectively. For each example, we compare our proposed methods with 1) Lasso: Lasso method which only uses the samples with complete observations; 2) Imputed-Lasso: Lasso method which uses all samples with missing data imputed by the Soft-thresholded SVD method (Mazumder et al. (2010)); 3) Ridge: Ridge regression method which only uses the samples with complete observations; 4) Imputed-Ridge: Ridge regression method which uses all samples with missing data imputed by the Soft-thresholded SVD method; and 5) IMSF (Yuan et al. (2012)): the IMSF method which uses all available data without imputing the missing data. We study four simulated examples, where the data are generated from the Gaussian distribution or some heavy-tailed distributions.
For each example, the data are generated from three modalities and each modality has 100 predictors. The training data set is composed of 100 samples with complete observations, 100 samples with observations from the first and the second modalities, 100 samples with observations from the first and the third modalities, and 100 samples with observations only from the first modality. The tuning data set contains 200 samples with complete observations and the testing data set contains 400 samples with complete observations. All methods use the tuning data set to choose the best tuning parameters. For the four simulated examples, samples with complete observations are generated from the linear model as follows.
Example 1:
The predictors with . The true coefficient vector
The true model is , where the errors .
Example 2:
The predictors , where Σ is a block diagonal matrix with p/5 blocks. Each block is a 5 × 5 square matrix with ones on the main diagonal and 0.15 elsewhere. The true coefficient vector
The true model is , where the errors .
Example 3:
The predictors , where Σ is the same as the covariance matrix shown in Example 1. For this multivariate t-distribution with the degrees of freedom 5, the variances of all predictors are equal to 1. The true coefficient vector β0 is the same as the vector shown in Example 1. The true model is , where the errors ϵ1, ϵ2, . . ., ϵn follow the Student’s t-distribution with degrees of freedom 10.
Example 4:
The predictors ∼ the mixture distribution , where ρ = 0.03 and I is a p × p identity matrix. The true coefficient vector β0 is the same as the vector shown in Example 1. The true model is , where the errors ϵ1, ϵ2, . . ., ϵn follow the Skew-t distribution (Azzalini (2013)) with degrees of freedom 4.
For each example, we repeated the simulation 30 times. To evaluate different methods, we use the following five measures: distance , mean squared error (MSE), false positive rate (FPR), false negative rate (FNR), and the elapsed time (in seconds) using R. Tables 1 and 2 show the performance comparison of different methods in the Gaussian case and the heavy-tailed case, respectively. The results indicate that our proposed methods deliver the best performance on all these four examples. For the Gaussian case shown in Table 1, DISCOM delivers better performance than the DISCOM-Huber method. For the heavy-tailed case shown in Table 2, DISCOM-Huber performs better. These numerical results are consistent with our theoretical studies shown in Section 3.
Table 1:
Performance comparison for the Gaussian case.
| Methods |
Example 1
|
Example 2
|
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| MSE | FPR | FNR | TIME | MSE | FPR | FNR | TIME | |||
| Lasso | 0.655 (0.026) |
1.431 (0.045) |
0.069 (0.004) |
0.015 (0.009) |
0.016 (0.000) |
0.920 (0.025) |
1.988 (0.059) |
0.133 (0.007) |
0.002 (0.002) |
0.019 (0.004) |
| Imputed-Lasso | 0.674 (0.017) |
1.338 (0.018) |
0.076 (0.007) |
0.004 (0.004) |
0.802 (0.006) |
0.690 (0.013) |
1.546 (0.030) |
0.122 (0.007) |
0.000 (0.000) |
1.099 (0.008) |
| Ridge | 1.270 (0.004) |
3.962 (0.062) |
1.000 (0.000) |
0.000 (0.000) |
0.025 (0.000) |
1.662 (0.006) |
5.262 (0.066) |
1.000 (0.000) |
0.000 (0.000) |
0.025 (0.000) |
| Imputed-Ridge | 1.094 (0.013) |
2.304 (0.035) |
1.000 (0.000) |
0.000 (0.000) |
0.780 (0.006) |
1.332 (0.009) |
3.130 (0.048) |
1.000 (0.000) |
0.000 (0.000) |
1.093 (0.008) |
| IMSF | 0.585 (0.020) |
1.358 (0.037) |
0.173 (0.009) |
0.000 (0.000) |
5.554 (0.068) |
0.777 (0.016) |
1.730 (0.040) |
0.291 (0.012) |
0.000 (0.000) |
5.900 (0.075) |
| DISCOM | 0.416 (0.013) |
1.133 (0.016) |
0.025 (0.003) |
0.000 (0.000) |
13.552 (0.078) |
0.600 (0.020) |
1.378 (0.033) |
0.074 (0.007) |
0.000 (0.000) |
12.391 (0.064) |
| DISCOM-Huber | 0.434 (0.013) |
1.145 (0.016) |
0.026 (0.003) |
0.000 (0.000) |
28.618 (0.886) |
0.605 (0.021) |
1.380 (0.035) |
0.076 (0.008) |
0.000 (0.000) |
25.907 0.122 |
| Fast-DISCOM | 0.465 (0.015) |
1.160 (0.016) |
0.039 (0.005) |
0.000 (0.000) |
3.600 (0.027) |
0.641 (0.017) |
1.438 (0.033) |
0.109 (0.006) |
0.000 (0.000) |
3.241 (0.029) |
| Fast-DISCOM-Huber | 0.481 (0.015) |
1.173 (0.016) |
0.036 (0.004) |
0.000 (0.000) |
16.802 (0.081) |
0.655 (0.020) |
1.457 (0.037) |
0.100 (0.007) |
0.000 (0.000) |
16.767 (0.096) |
[Note that the values in the parentheses are the standard errors of the measures.]
Table 2:
Performance comparison for the heavy-tailed case.
| Methods |
Example 3
|
Example 4
|
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| MSE | FPR | FNR | TIME | MSE | FPR | FNR | TIME | |||
| Lasso | 0.751 (0.036) |
1.809 (0.055) |
0.070 (0.006) |
0.056 (0.017) |
0.021 (0.004) |
1.305 (0.029) |
3.331 (0.087) |
0.064 (0.007) |
0.419 (0.054) |
0.020 (0.005) |
| Imputed-Lasso | 0.751 (0.023) |
1.669 (0.039) |
0.071 (0.008) |
0.026 (0.010) |
0.687 (0.010) |
0.930 (0.030) |
2.699 (0.073) |
0.147 (0.014) |
0.033 (0.016) |
0.530 (0.013) |
| Ridge | 1.294 (0.004) |
4.454 (0.114) |
1.000 (0.000) |
0.000 (0.000) |
0.028 (0.003) |
1.420 (0.006) |
3.548 (0.069) |
1.000 (0.000) |
0.000 (0.000) |
0.039 (0.006) |
| Imputed-Ridge | 1.143 (0.013) |
2.731 (0.064) |
1.000 (0.000) |
0.000 (0.000) |
0.657 (0.010) |
1.326 (0.011) |
3.342 (0.080) |
1.000 (0.000) |
0.000 (0.000) |
0.527 (0.011) |
| IMSF | 0.622 (0.025) |
1.637 (0.041) |
0.173 (0.013) |
0.004 (0.004) |
6.569 (0.297) |
1.048 (0.028) |
2.878 (0.083) |
0.189 (0.012) |
0.052 (0.017) |
6.989 (0.188) |
| DISCOM | 0.579 (0.022) |
1.560 (0.038) |
0.037 (0.004) |
0.004 (0.004) |
12.086 (0.134) |
0.871 (0.025) |
2.590 (0.067) |
0.193 (0.017) |
0.011 (0.006) |
12.362 (0.153) |
| DISCOM-Huber | 0.507 (0.017) |
1.452 (0.025) |
0.027 (0.003) |
0.000 (0.000) |
26.073 (0.104) |
0.780 (0.021) |
2.468 (0.054) |
0.137 (0.012) |
0.004 (0.004) |
26.925 (0.228) |
| Fast-DISCOM | 0.601 (0.022) |
1.604 (0.047) |
0.040 (0.004) |
0.004 (0.004) |
3.317 (0.041) |
1.151 (0.025) |
3.028 (0.079) |
0.207 (0.019) |
0.085 (0.033) |
3.626 (0.050) |
| Fast-DISCOM-Huber | 0.561 (0.021) |
1.496 (0.031) |
0.035 (0.004) |
0.000 (0.000) |
17.835 (0.079) |
0.786 (0.022) |
2.482 (0.055) |
0.137 (0.013) |
0.000 (0.000) |
17.042 (0.134) |
[Note that the values in the parentheses are the standard errors of the measures.]
In addition, as shown in Tables 1 and 2, for the Lasso and ridge regression, using the imputed data can improve performance in most cases. However, as shown in Table 1, the Lasso method using the imputed data may deliver worse estimate of the true coefficient vector β0, possibly due to the block-missing pattern. Compared with the Lasso and Ridge regression methods using the imputed data set or only the samples with complete observations, the IMSF method delivers better estimation and prediction. On the other hand, IMSF method has high false positive rates for all four simulated examples. The comparison between IMSF and our proposed DISCOM and DISCOM-Huber shows that our proposed methods could use all available data more effectively and therefore acquires better performance.
For each simulation of the four examples, our proposed Fast-DISCOM method using the fast tuning parameter selection method uses only 4 seconds while our original DISCOM method uses about 13 seconds. The Fast-DISCOM method is also faster than the IMSF method which uses about 7 seconds for each simulation. On the other hand, we can observe that the computing times of our original DISCOM and DISCOM-Huber methods are still acceptable. For the examples 1 and 2 generated from the Gaussian distribution, although the Fast-DISCOM method does not perform as well as the DISCOM method, it has better estimation, prediction, and model selection performance than the Lasso, ridge regression and IMSF methods. Similarly, for the examples 3 and 4 generated from the heavy-tailed distributions, although the Fast-DISCOM-Huber method does not perform as well as the DISCOM-Huber method, it also has better performance than the Lasso, ridge regression and IMSF methods. These simulation results indicate that our proposed new tuning parameter selection method accelerates the computational speed without sacrificing the estimation, prediction, and model selection performance too much.
5. Real Data Analysis
In this section, we show the analysis of the Alzheimer’s Disease Neuroimaging Initiative (ADNI) data as an application example. The main goal of ADNI is to test whether serial magnetic resonance imaging (MRI), positron emission tomography (PET), and some other biological markers and neuropsychological assessments can be combined to measure the progression of mild cognitive impairment (MCI) and early Alzheimer’s disease (AD). In our study, we extracted features from three modalities: structural MRI, fluorodeoxyglucose PET, and CerebroSpinal Fluid (CSF). Imaging preprocessing was performed for MRI and PET images. For the MRI, after some correction, spatial segmentation, and registration steps, we obtained the subject lableled image based on the Jacob template (Kabani et al. (1998)) with 93 manually labeled regions of interest (ROI). For each of the 93 ROIs in the labeled MRI, we computed the volume of gray matter as a feature. For each PET image, we first aligned the PET image to its respective MRI using affine registration. Then, we calculated the average intensity of every ROI in the PET image as a feature. Therefore, for each ROI, we have one MRI feature and one PET feature. For the CSF modality, five biomarkers were used in this study, namely amyloid β (Aβ42), CSF total tau (t-tau), tau hyperphosphorylated at threonine 181 (p-tau), and two tau ratios with respective to Aβ42 (i.e., t-tau/Aβ42 and p-tau/Aβ42).
After data processing, we got 93 features from MRI, 93 features from PET, and 5 features from CSF. There are 805 subjects in total, including 1) 199 subjects with complete MRI, PET, and CSF features, 2) 197 subjects with only MRI and PET features, 3) 201 subjects with only MRI and CSF features, and 4) 208 subjects with only MRI features. The response variable used in our study is the Mini Mental State Examination (MMSE) score. As a brief 30-point questionnaire test, MMSE can be used to examine a patient’s arithmetic, memory and orientation (Folstein et al. (1975)). It is very useful to help evaluate the stage of AD pathology and predict future progression. We will use all available data from MRI, PET, and CSF to predict the MMSE score.
In our analysis, we divided the data into three parts: training data set, tuning data set, and testing data set. The training data set consists of all subjects with incomplete observations and 40 randomly selected subjects with complete MRI, PET, and CSF features. The tuning data set consists of another 40 randomly selected subjects (different from the training data set) with complete observations. The testing data set contains the other 119 subjects with complete observations. The tuning data set was used to choose the best tuning parameters for all methods and the testing data set was used to evaluate different methods. We used all methods shown in the simulation study to predict the MMSE score. For each method, the analysis was repeated 30 times using different partitions of the data.
The results in Table 3 show that our proposed Fast-DISCOM-Huber method acquires the best prediction performance. All our proposed DISCOM methods deliver better performance than the Lasso, Ridge, and IMSF methods. The IMSF method has better prediction performance than the Lasso and ridge regression using only samples with complete observations. However, IMSF does not perform as well as the ridge regression using the imputed data. Regarding the model selection, since the number of variables selected by the Lasso is at most the sample size (Zou and Hastie (2005)), as shown in Table 3, the Lasso method using the imputed data selected many more features than the method using only samples with complete observations. Both IMSF and our proposed methods could deliver a model with relatively small numbers of features.
Table 3:
Performance comparison for the ADNI data.
| Methods | MSE |
Number of Features |
TIME |
|||
|---|---|---|---|---|---|---|
| Mean | SE | Mean | SE | Mean | SE | |
| Lasso | 5.711 | 0.341 | 11.733 | 1.638 | 0.009 | 0.002 |
| Imputed-Lasso | 4.711 | 0.082 | 86.700 | 8.559 | 0.559 | 0.017 |
| Ridge | 5.273 | 0.204 | 191.000 | 0.000 | 0.010 | 0.000 |
| Imputed-Ridge | 4.478 | 0.055 | 191.000 | 0.000 | 0.177 | 0.006 |
| IMSF | 4.630 | 0.079 | 28.400 | 3.025 | 2.960 | 0.073 |
| DISCOM | 4.285 | 0.068 | 27.933 | 2.261 | 4.675 | 0.028 |
| DISCOM-Huber | 4.161 | 0.059 | 23.100 | 0.846 | 10.348 | 0.025 |
| Fast-DISCOM | 4.146 | 0.055 | 28.100 | 0.809 | 1.565 | 0.007 |
| Fast-DISCOM-Huber | 4.123 | 0.069 | 25.833 | 1.311 | 8.012 | 0.019 |
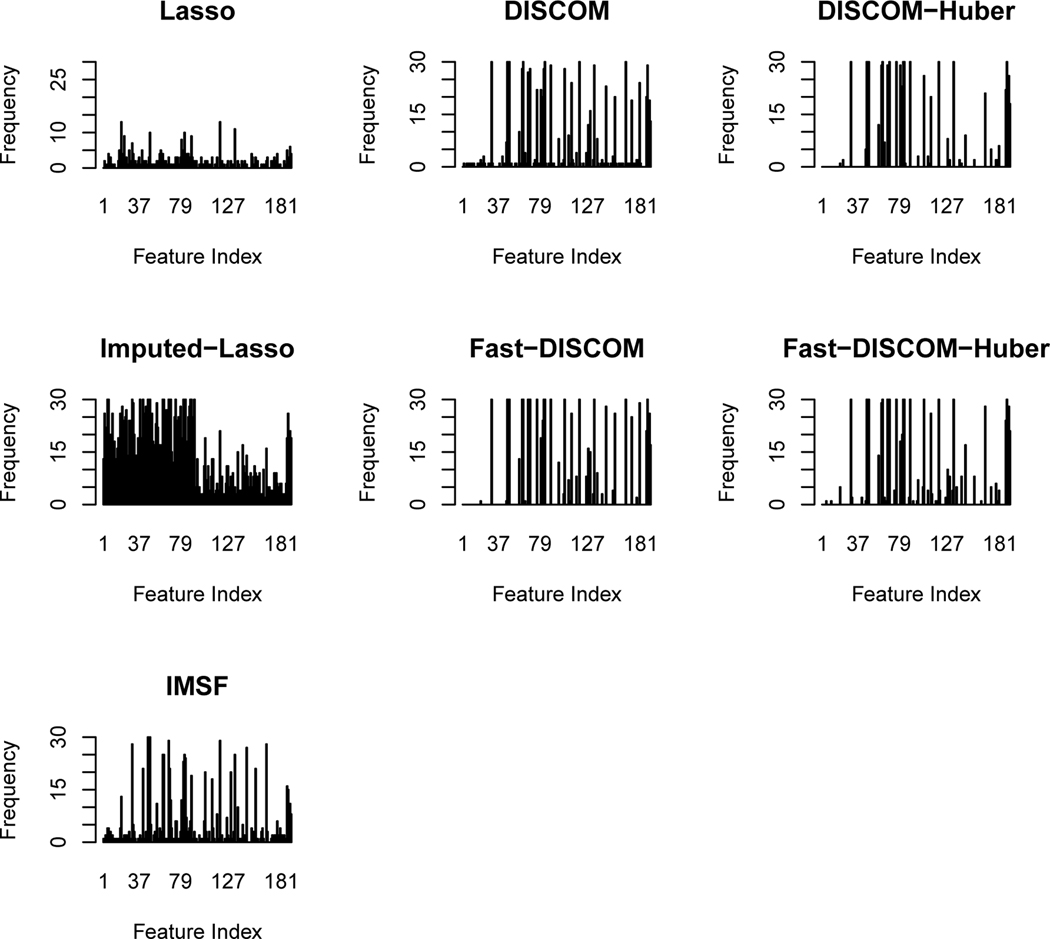
Figure 2 shows the selection frequency of all the 191 features. The selection frequency of each feature is defined as the times of being selected in the 30 times replications. As shown in Figure 2, for our proposed DISCOM methods, some features were always selected and many features were never selected in the 30 times replications. This means that our method could deliver relatively robust performance on model selection. However, for some other methods such as the Imputed-Lasso method, they selected very different features in different replications and therefore many features have positive and low selection frequencies. For the Imputed-Lasso method, one possible reason for the unstable performance on model selection is due to the randomness involved in the imputation of a lot of block-missing data.
Figure 2:
Selection frequency of 191 features for the prediction of MMSE score.
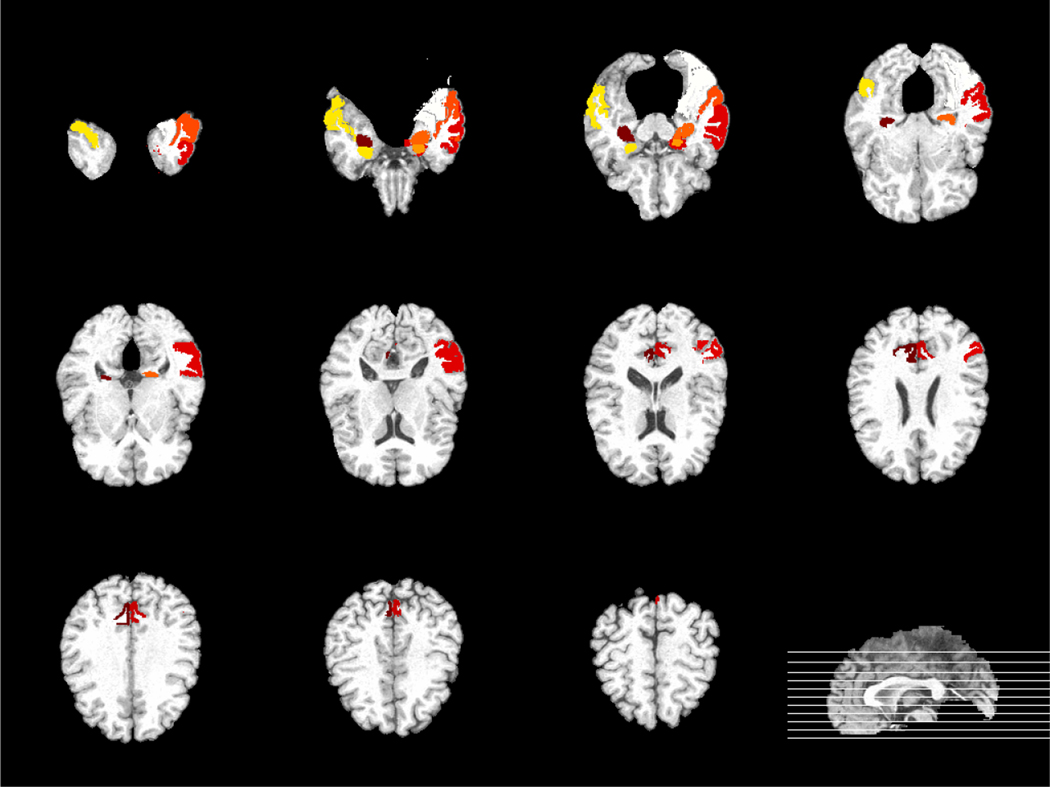
To further understand our results, since each MRI feature and each PET feature are corresponding to one ROI, we can examine whether the selected features are meaningful by studying their corresponding brain regions. In our 30 times of experiments using different random splits, there are 9 MRI features and 2 PET features always selected by our proposed DISCOM-Huber and Fast-DISCOM-Huber methods. Figure 3 shows the multi-slice view of the brain regions (regions with color) corresponding to these 11 features. Among these 11 brain regions, some regions such as hippocampal formation right (30-th region), uncus left (46-th region), middle temporal gyrus left (48-th region), hippocampus formation left (69-th region) and amygdale right (83-th region), are known to be highly correlated with AD and MCI by many studies using group comparison methods (Misra et al. (2009); Zhang and Shen (2012)). It would be interesting to study whether the other six always selected brain regions are truly related with AD by some scientific experiments.
Figure 3:
The multi-slice view of the brain regions always selected by DISCOM-Huber and Fast-DISCOM-Huber.
In addition, as shown in Table 3, all our proposed DISCOM methods solve this real data analysis problem with 191 features within 11 seconds. This indicates that the time cost of our methods is not very expensive. In summary, our real data analysis indicates that our proposed method can solve practical problems well.
6. Conclusion
In this paper, we propose a new two-step procedure to find the optimal linear prediction of a continuous response variable using the block-missing multi-modality predictors. In the first step, we estimate the covariance matrix of the predictors using a linear combination of the identity matrix, and the estimates of the intra-modality covariance matrix and the cross-modality covariance matrix. The proposed estimator of the covariance matrix can be positive semi-definite and more accurate than the sample covariance matrix. We also use all available information to estimate the cross covariance vector between the predictors and the response variable. Robust estimate based on the Huber’s M-estimate is also proposed for the heavy-tailed case. In the second step, based on the estimated covariance matrix and the cross-covariance vector, a modified Lasso estimator is used to deliver a sparse estimate of the coefficients in the optimal linear prediction. The effectiveness of the proposed method is demonstrated by both theoretical and numerical studies. The comparison between our proposed method and several existing ones also indicates that our method has promising performance on estimation, prediction, and model selection for the block-missing multi-modality data.
Supplementary Material
Acknowledgements
The authors would like to thank the editor, the associate editor, and reviewers, whose helpful comments and suggestions led to a much improved presentation. This research was partially supported by NSF grants IIS1632951, DMS1821231 and NIH grants R01GM126550, P01CA142538.
Contributor Information
Guan Yu, Department of Biostatistics, State University of New York at Buffalo.
Quefeng Li, Department of Biostatistics, University of North Carolina at Chapel Hill.
Dinggang Shen, Department of Radiology and BRIC, University of North Carolina at Chapel Hill. He is also affiliated with the Department of Brain and Cognitive Engineering, Korea University, Seoul 136-701, Korea.
Yufeng Liu, Department of Statistics and Operations Research, Department of Genetics, Department of Biostatistics, Carolina Center for Genome Science, Lineberger Comprehensive Cancer Center, University of North Carolina at Chapel Hill, NC 27599.
References
- Azzalini A (2013), The skew-normal and related families, vol. 3, Cambridge University Press. [Google Scholar]
- Bickel PJ and Levina E (2008), “Covariance regularization by thresholding,” The Annals of Statistics, 2577–2604.
- Bühlmann P and Van De Geer S (2011), Statistics for high-dimensional data: methods, theory and applications, Springer Science & Business Media. [Google Scholar]
- Bunea F et al. (2008), “Consistent selection via the Lasso for high dimensional approximating regression models,” in Pushing the limits of contemporary statistics: contributions in honor of Jayanta K. Ghosh, Institute of Mathematical Statistics, pp. 122–137. [Google Scholar]
- Cai J, Candès EJ, and Shen Z (2010), “A singular value thresholding algorithm for matrix completion,” SIAM Journal on Optimization, 20, 1956–1982. [Google Scholar]
- Cai T, Cai TT, and Zhang A (2016), “Structured matrix completion with applications to genomic data integration,” Journal of the American Statistical Association, 111, 621–633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai T and Liu W (2011), “Adaptive thresholding for sparse covariance matrix estimation,” Journal of the American Statistical Association, 106, 672–684. [Google Scholar]
- Cai TT and Zhang A (2016), “Minimax rate-optimal estimation of high-dimensional covariance matrices with incomplete data,” Journal of Multivariate Analysis, 150, 55–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catoni O (2012), “Challenging the empirical mean and empirical variance: a deviation study,” in Annales de l’Institut Henri Poincaré, Probabilités et Statistiques, Institut Henri Poincaré, vol. 48, pp. 1148–1185. [Google Scholar]
- Datta A, Zou H, et al. (2017), “Cocolasso for high-dimensional error-in-variables regression,” The Annals of Statistics, 45, 2400–2426. [Google Scholar]
- Fan J, Li Q, and Wang Y (2016), “Estimation of high dimensional mean regression in the absence of symmetry and light tail assumptions,” Journal of the Royal Statistical Society: Series B (Statistical Methodology), to appear. [DOI] [PMC free article] [PubMed]
- Fan J and Li R (2001), “Variable selection via nonconcave penalized likelihood and its oracle properties,” Journal of the American Statistical Association, 96, 1348–1361. [Google Scholar]
- Folstein MF, Folstein SE, and McHugh PR (1975), “Mini-mental state: a practical method for grading the cognitive state of patients for the clinician,” Journal of Psychiatric Research, 12, 189–198. [DOI] [PubMed] [Google Scholar]
- Friedman J, Hastie T, and Tibshirani R (2010), “Regularization paths for generalized linear models via coordinate descent,” Journal of Statistical Software, 33, 1–22. [PMC free article] [PubMed] [Google Scholar]
- Hastie T, Tibshirani R, Sherlock G, Eisen M, Brown P, and Botstein D (1999), “Imputing missing data for gene expression arrays,” Tech. rep, Stanford University. [Google Scholar]
- Huber PJ et al. (1964), “Robust estimation of a location parameter,” The Annals of Mathematical Statistics, 35, 73–101. [Google Scholar]
- Jeng XJ and Daye ZJ (2011), “Sparse covariance thresholding for high-dimensional variable selection,” Statistica Sinica, 625–657.
- Kabani N, MacDonald D, Holmes C, and Evans A (1998), “A 3D atlas of the human brain,” NeuroImage, 7, S717. [Google Scholar]
- Ledoit O and Wolf M (2004), “A well-conditioned estimator for large-dimensional covariance matrices,” Journal of Multivariate Analysis, 88, 365–411. [Google Scholar]
- Loh P-L and Wainwright MJ (2012), “High-dimensional regression with noisy and missing data: provable guarantees with nonconvexity,” The Annals of Statistics, 40, 1637–1664. [Google Scholar]
- Lounici K et al. (2014), “High-dimensional covariance matrix estimation with missing observations,” Bernoulli, 20, 1029–1058. [Google Scholar]
- Mazumder R, Hastie T, and Tibshirani R (2010), “Spectral regularization algorithms for learning large incomplete matrices,” The Journal of Machine Learning Research, 11, 2287–2322. [PMC free article] [PubMed] [Google Scholar]
- Meinshausen N and Bühlmann P (2006), “High-dimensional graphs and variable selection with the lasso,” The Annals of Statistics, 1436–1462.
- Meinshausen N and Yu B (2009), “Lasso-type recovery of sparse representations for high-dimensional data,” The Annals of Statistics, 246–270.
- Misra C, Fan Y, and Davatzikos C (2009), “Baseline and longitudinal patterns of brain atrophy in MCI patients, and their use in prediction of short-term conversion to AD: results from ADNI,” NeuroImage, 44, 1415–1422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raskutti G, Wainwright MJ, and Yu B (2010), “Restricted eigenvalue properties for correlated Gaussian designs,” Journal of Machine Learning Research, 11, 2241–2259. [Google Scholar]
- Raskutti G, Wainwright MJ, and Yu B (2011), “Minimax rates of estimation for high-dimensional linear regression over ℓq-balls,” IEEE Transactions on Information Theory, 57, 6976–6994. [Google Scholar]
- Ravikumar P, Wainwright MJ, Raskutti G, and Yu B (2011), “High-dimensional covariance estimation by minimizing ℓ1-penalized log-determinant divergence,” Electronic Journal of Statistics, 5, 935–980. [Google Scholar]
- Rothman AJ (2012), “Positive definite estimators of large covariance matrices,” Biometrika, 99, 733–740. [Google Scholar]
- Tibshirani R (1996), “Regression shrinkage and selection via the lasso,” Journal of the Royal Statistical Society: Series B, 58, 267–288. [Google Scholar]
- Van De Geer SA, Bühlmann P, et al. (2009), “On the conditions used to prove oracle results for the Lasso,” Electronic Journal of Statistics, 3, 1360–1392. [Google Scholar]
- Wainwright MJ (2009), “Sharp thresholds for High-Dimensional and noisy sparsity recovery using ℓ1-Constrained Quadratic Programming (Lasso),” IEEE Transactions on Information Theory, 55, 2183–2202. [Google Scholar]
- Yuan L, Wang Y, Thompson PM, Narayan VA, and Ye J (2012), “Multi-source feature learning for joint analysis of incomplete multiple heterogeneous neuroimaging data,” NeuroImage, 61, 622–632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C-H (2010), “Nearly unbiased variable selection under minimax concave penalty,” The Annals of Statistics, 38, 894–942. [Google Scholar]
- Zhang C-H, Huang J, et al. (2008), “The sparsity and bias of the lasso selection in high-dimensional linear regression,” The Annals of Statistics, 36, 1567–1594. [Google Scholar]
- Zhang D and Shen D (2012), “Multi-modal multi-task learning for joint prediction of multiple regression and classification variables in Alzheimer’s disease,” NeuroImage, 59, 895–907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao P and Yu B (2006), “On model selection consistency of Lasso,” The Journal of Machine Learning Research, 7, 2541–2563. [Google Scholar]
- Zhou S, van de Geer S, and Bühlmann P (2009), “Adaptive Lasso for high dimensional regression and Gaussian graphical modeling,” arXiv preprint arXiv:0903.2515.
- Zou H (2006), “The Adaptive Lasso and Its Oracle Properties,” Journal of the American Statistical Association, 101, 1418–1429. [Google Scholar]
- Zou H and Hastie T (2005), “Regularization and variable selection via the elastic net,” Journal of the Royal Statistical Society: Series B, 67, 301–320. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.