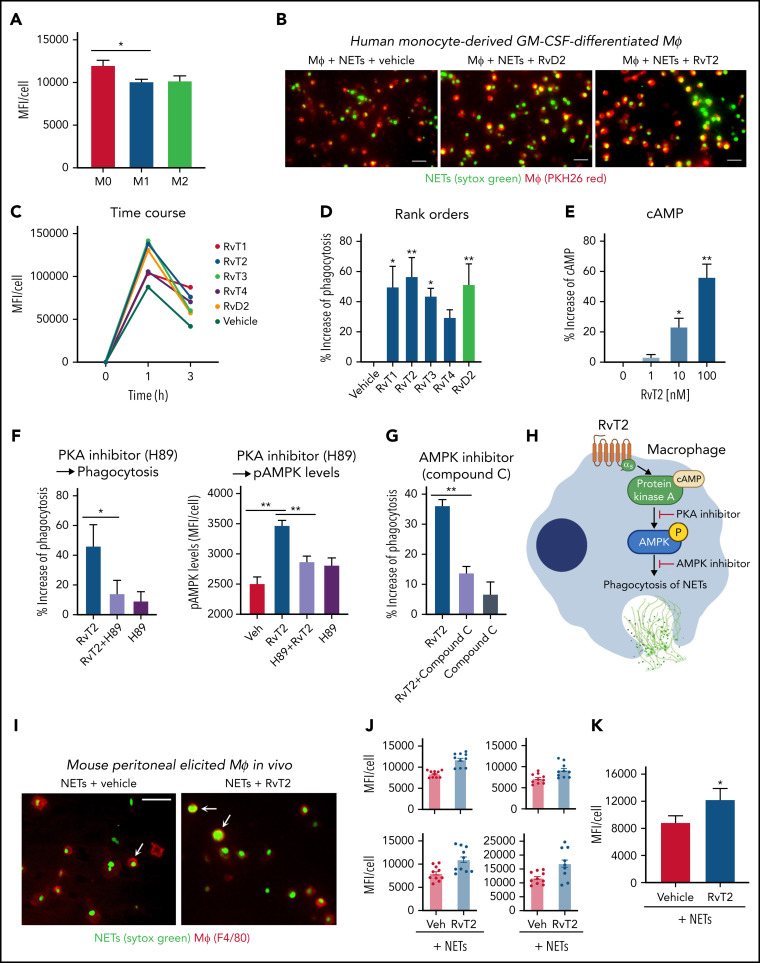
Figure 5.
RvTs enhance MΦ ingestion of NET in vitro and in vivo. (A-D) Human MΦs were plated onto 8-well chamber slides (1 × 105 cells per well). Test compounds (RvT1-RvT4, RvD2; 10 nM) or vehicle controls were added to MΦs for 15 minutes, followed by the addition of Sytox Green–labeled NETs for 1 hour. NETs from ∼1 × 106 PMNs (∼10 μg DNA) were added to 1 × 105 MΦs per well. (A) Mean fluorescence intensity (MFI) per cell with M0-MΦ, M1-MΦ, and M2-MΦ. Mean ± SEM. n = 3 separate donors. *P < .05. (B) Representative images of M0-MΦs (PKH26 Red) with ingested Sytox Green–labeled NETs. Scale bars = 50 μm. (C-D) An average of ∼300 MΦs were quantified in each condition for each donor. (C) Time course of MFI per MΦ: a representative donor from n = 4 separate donors. (D) Results are percent increase of MFI/MΦ compared with MΦ + NET + vehicle. Mean ± SEM. n = 8 separate donors. *P < .05, **P < .01, one-way analysis of variance with Tukey's multiple comparisons. (E) cAMP. Human M0-MΦs (1 × 105/0.2 mL) were incubated with RvT2 (0-100 nM) for 15 minutes. MΦs were lysed and cAMP levels determined. Results are percent increase above vehicle. Mean ± SEM. n = 3. *P < .05, **P < .01, vs vehicle. (F-G) Human M0-MΦs (1 × 105 cells/well in 8-well chamber slides) were incubated with RvT2 (10 nM), a PKA inhibitor (H89, 3 μM, 24 hours), the AMPK inhibitor (Compound C, 500 nM, 4 hours), or vehicle controls for 15 minutes, followed by the addition of Sytox Green–labeled NETs for 1 hour. MΦ ingestion of NETs was quantified according to Sytox Green intensities and pAMPK levels quantified according to immunofluorescence with an anti–phospho-AMPK (pAMPK) antibody. Mean ± SEM. n = 5 (phagocytosis) or n = 3 (pAMPK levels) separate donors. *P < .05, **P < .01. (H) Proposed RvT2-cAMP-PKA-AMPK axis in stimulating MΦ phagocytosis of NETs. (I-K) Mice were given zymosan (1 mg/mL, IP) for 72 hours, followed by RvT2 (100 ng/mL per mouse, IP) or vehicle (0.05% ethanol) for 15 minutes, and fluorescent-labeled NETs (∼100 μg/mL DNA from ∼10 × 106 PMN) for 60 minutes. Exudate cells (2 × 105 cells) were adhered onto 8-well chamber slides for imaging and quantification. An average of ∼780 MΦs was quantified per condition per mouse. (I) Representative images of MΦs (red: F4/80) with ingested Sytox Green–labeled NETs, denoted by arrows. Scale bars = 50 μm. (J) Four independent experiments and 4 mice each group. In each experiment, 10 fields (20×) per mouse were imaged and quantified. (K) Mean ± SEM from n = 4. *P < .05, two-tailed paired Student t test. GM-CSF, granulocyte-macrophage colony-stimulating factor; Veh, vehicle control.